Biomedical diagnosis perspective of epigenetic detections using alpha-hemolysin nanopore
- PMID: 30931380
- PMCID: PMC6436813
- DOI: 10.3934/matersci.2015.4.448
Biomedical diagnosis perspective of epigenetic detections using alpha-hemolysin nanopore
Abstract
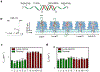
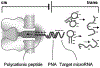
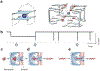
The α-hemolysin nanopore has been studied for applications in DNA sequencing, various single-molecule detections, biomolecular interactions, and biochips. The detection of single molecules in a clinical setting could dramatically improve cancer detection and diagnosis as well as develop personalized medicine practices for patients. This brief review shortly presents the current solid state and protein nanopore platforms and their applications like biosensing and sequencing. We then elaborate on various epigenetic detections (like microRNA, G-quadruplex, DNA damages, DNA modifications) with the most widely used alpha-hemolysin pore from a biomedical diagnosis perspective. In these detections, a nanopore electrical current signature was generated by the interaction of a target with the pore. The signature often was evidenced by the difference in the event duration, current level, or both of them. An ideal signature would provide obvious differences in the nanopore signals between the target and the background molecules. The development of cancer biomarker detection techniques and nanopore devices have the potential to advance clinical research and resolve health problems. However, several challenges arise in applying nanopore devices to clinical studies, including super low physiological concentrations of biomarkers resulting in low sensitivity, complex biological sample contents resulting in false signals, and fast translocating speed through the pore resulting in poor detections. These issues and possible solutions are discussed.
Keywords: DNA damage; DNA hydroxymethylation; DNA methylation; G-quadruplex; abasic site; cancer biomarker; guanine oxidation; microRNA; nanopore device; α-hemolysin.
Conflict of interest statement
Conflict of Interest All authors declare no conflict of interest.
Figures
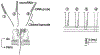




Similar articles
-
Characterization of Interstrand DNA-DNA Cross-Links Using the α-Hemolysin Protein Nanopore.ACS Nano. 2015 Dec 22;9(12):11812-9. doi: 10.1021/acsnano.5b03923. Epub 2015 Nov 18. ACS Nano. 2015. PMID: 26563913 Free PMC article.
-
Biological Nanopores: Confined Spaces for Electrochemical Single-Molecule Analysis.Acc Chem Res. 2018 Feb 20;51(2):331-341. doi: 10.1021/acs.accounts.7b00143. Epub 2018 Jan 24. Acc Chem Res. 2018. PMID: 29364650
-
Insights into protein sequencing with an α-Hemolysin nanopore by atomistic simulations.Sci Rep. 2019 Apr 23;9(1):6440. doi: 10.1038/s41598-019-42867-7. Sci Rep. 2019. PMID: 31015503 Free PMC article.
-
Glass Capillary-Based Nanopores for Single Molecule/Single Cell Detection.ACS Sens. 2023 Feb 24;8(2):427-442. doi: 10.1021/acssensors.2c02102. Epub 2023 Jan 20. ACS Sens. 2023. PMID: 36670058 Review.
-
Single molecule sensing by nanopores and nanopore devices.Analyst. 2010 Mar;135(3):441-51. doi: 10.1039/b907735a. Epub 2009 Dec 22. Analyst. 2010. PMID: 20174694 Free PMC article. Review.
Cited by
-
Single-molecule analysis of interaction between p53TAD and MDM2 using aerolysin nanopores.Chem Sci. 2021 Mar 25;12(16):5883-5891. doi: 10.1039/d1sc00386k. Chem Sci. 2021. PMID: 34168813 Free PMC article.
-
A Protein Nanopore-Based Approach for Bacteria Sensing.Nanoscale Res Lett. 2016 Dec;11(1):501. doi: 10.1186/s11671-016-1715-z. Epub 2016 Nov 15. Nanoscale Res Lett. 2016. PMID: 27848237 Free PMC article.
-
A Solid-State Hard Microfluidic-Nanopore Biosensor with Multilayer Fluidics and On-Chip Bioassay/Purification Chamber.Adv Funct Mater. 2018 Dec 12;28(50):1804182. doi: 10.1002/adfm.201804182. Epub 2018 Oct 16. Adv Funct Mater. 2018. PMID: 31632230 Free PMC article.
-
Nanoscale Investigation of Generation 1 PAMAM Dendrimers Interaction with a Protein Nanopore.Sci Rep. 2017 Jul 21;7(1):6167. doi: 10.1038/s41598-017-06435-1. Sci Rep. 2017. PMID: 28733599 Free PMC article.
-
Recent advances in biological nanopores for nanopore sequencing, sensing and comparison of functional variations in MspA mutants.RSC Adv. 2021 Aug 31;11(46):28996-29014. doi: 10.1039/d1ra02364k. eCollection 2021 Aug 23. RSC Adv. 2021. PMID: 35478559 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
