Structure-Based Design, Synthesis, and Biological Evaluation of Non-Acyl Sulfamate Inhibitors of the Adenylate-Forming Enzyme MenE
- PMID: 30912442
- PMCID: PMC6653581
- DOI: 10.1021/acs.biochem.9b00003
Structure-Based Design, Synthesis, and Biological Evaluation of Non-Acyl Sulfamate Inhibitors of the Adenylate-Forming Enzyme MenE
Abstract
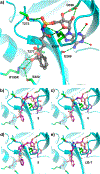
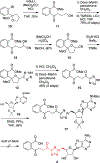
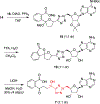
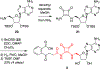
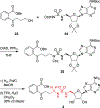
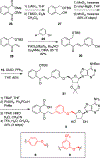
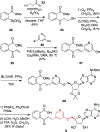
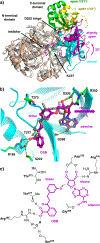
N-Acyl sulfamoyladenosines (acyl-AMS) have been used extensively to inhibit adenylate-forming enzymes that are involved in a wide range of biological processes. These acyl-AMS inhibitors are nonhydrolyzable mimics of the cognate acyl adenylate intermediates that are bound tightly by adenylate-forming enzymes. However, the anionic acyl sulfamate moiety presents a pharmacological liability that may be detrimental to cell permeability and pharmacokinetic profiles. We have previously developed the acyl sulfamate OSB-AMS (1) as a potent inhibitor of the adenylate-forming enzyme MenE, an o-succinylbenzoate-CoA (OSB-CoA) synthetase that is required for bacterial menaquinone biosynthesis. Herein, we report the use of computational docking to develop novel, non-acyl sulfamate inhibitors of MenE. A m-phenyl ether-linked analogue (5) was found to be the most potent inhibitor (IC50 = 8 μM; Kd = 244 nM), and its X-ray co-crystal structure was determined to characterize its binding mode in comparison to the computational prediction. This work provides a framework for the development of potent non-acyl sulfamate inhibitors of other adenylate-forming enzymes in the future.
Conflict of interest statement
The authors declare the following competing financial interest(s): D.S.T., P.J.T., C.E.E., and J.S.M. are co-inventors on International Patent Application PCT/US2016/055136; D.S.T., P.J.T., C.E.E., and Y.S. are co-inventors on U.S. Provisional Patent Application 62/802,650.
Figures
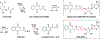
Similar articles
-
Mechanism of MenE inhibition by acyl-adenylate analogues and discovery of novel antibacterial agents.Biochemistry. 2015 Oct 27;54(42):6514-6524. doi: 10.1021/acs.biochem.5b00966. Epub 2015 Oct 15. Biochemistry. 2015. PMID: 26394156 Free PMC article.
-
Stable analogues of OSB-AMP: potent inhibitors of MenE, the o-succinylbenzoate-CoA synthetase from bacterial menaquinone biosynthesis.Chembiochem. 2012 Jan 2;13(1):129-36. doi: 10.1002/cbic.201100585. Epub 2011 Nov 23. Chembiochem. 2012. PMID: 22109989 Free PMC article.
-
Mechanistic Insights from the Crystal Structure of Bacillus subtilis o-Succinylbenzoyl-CoA Synthetase Complexed with the Adenylate Intermediate.Biochemistry. 2016 Dec 6;55(48):6685-6695. doi: 10.1021/acs.biochem.6b00889. Epub 2016 Nov 28. Biochemistry. 2016. PMID: 27933791
-
Targeting adenylate-forming enzymes with designed sulfonyladenosine inhibitors.J Antibiot (Tokyo). 2019 Jun;72(6):325-349. doi: 10.1038/s41429-019-0171-2. Epub 2019 Apr 15. J Antibiot (Tokyo). 2019. PMID: 30982830 Free PMC article. Review.
-
Sulfamates and their therapeutic potential.Med Res Rev. 2005 Mar;25(2):186-228. doi: 10.1002/med.20021. Med Res Rev. 2005. PMID: 15478125 Review.
Cited by
-
Structural approaches to pathway-specific antimicrobial agents.Transl Res. 2020 Jun;220:114-121. doi: 10.1016/j.trsl.2020.02.001. Epub 2020 Feb 6. Transl Res. 2020. PMID: 32105648 Free PMC article. Review.
-
TBPEH-TBPB Initiate the Radical Addition of Benzaldehyde and Allyl Esters.Int J Mol Sci. 2022 Nov 8;23(22):13704. doi: 10.3390/ijms232213704. Int J Mol Sci. 2022. PMID: 36430186 Free PMC article.
References
-
- Brown ED, and Wright GD (2016) Antibacterial drug discovery in the resistance era. Nature (London, U. K.) 529, 336–343. - PubMed
-
- Lewis K (2013) Platforms for antibiotic discovery. Nat. Rev. Drug Discovery 12, 371–387. - PubMed
-
- Centers for Disease Control and Prevention (2013) Antibiotic Resistance Threats in the United States, 2013 http://www.cdc.gov/drugresistance/threat-report-2013/.
-
- World Health Organization (2014) Antimicrobial resistance: Global report on surveillance 2014 http://www.who.int/drugresistance/documents/surveillancereport/en/.
-
- The Pew Charitable Trusts (2016) A Scientific Roadmap for Antibiotic Discovery https://www.pewtrusts.org/en/research-and-analysis/reports/2016/05/a-sci....
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

