A unique cluster of roo insertions in the promoter region of a stress response gene in Drosophila melanogaster
- PMID: 30911338
- PMCID: PMC6415491
- DOI: 10.1186/s13100-019-0152-9
A unique cluster of roo insertions in the promoter region of a stress response gene in Drosophila melanogaster
Abstract
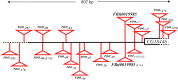
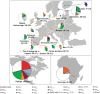
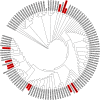
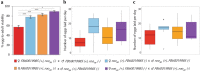
Transposable elements (TEs) are not randomly distributed in the genome. A genome-wide analysis of the D. melanogaster genome found that differences in TE density across 50 kb genomic regions was due both to transposition and duplication. At smaller genomic scales, promoter regions of hsp genes and the promoter region of CG18446 have been shown to accumulate TE insertions. In this work, we have further analyzed the promoter region of CG18446. We screened 218 strains collected in 15 natural populations, and we found that the CG18446 promoter region contains 20 independent roo insertions. Based on phylogenetic analysis, we suggest that the presence of multiple roo insertions in this region is likely to be the result of several bursts of transposition. Moreover, we found that the roo insertional cluster in the CG18446 promoter region is unique: no other promoter region in the genome contains a similar number of roo insertions. We found that, similar to hsp gene promoters, chromatin accessibility could be one of the factors explaining the recurrent insertions of roo elements in CG18446 promoter region.
Keywords: Fecundity; Natural population; Recurrent insertion; Target site duplication; Transposable element; Viability.
Conflict of interest statement
Not applicable.Not applicable.The authors declare that they have no competing interests.Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Multiple Independent Retroelement Insertions in the Promoter of a Stress Response Gene Have Variable Molecular and Functional Effects in Drosophila.PLoS Genet. 2016 Aug 12;12(8):e1006249. doi: 10.1371/journal.pgen.1006249. eCollection 2016 Aug. PLoS Genet. 2016. PMID: 27517860 Free PMC article.
-
Contrasting patterns of transposable element insertions in Drosophila heat-shock promoters.PLoS One. 2009 Dec 29;4(12):e8486. doi: 10.1371/journal.pone.0008486. PLoS One. 2009. PMID: 20041194 Free PMC article.
-
Long-term evolution of the roo transposable element copy number in mutation accumulation lines of Drosophila melanogaster.Genet Res (Camb). 2011 Jun;93(3):181-7. doi: 10.1017/S0016672311000103. Epub 2011 May 6. Genet Res (Camb). 2011. PMID: 21554776
-
Accumulation of transposable elements in laboratory lines of Drosophila melanogaster.Genetica. 1997;100(1-3):167-75. Genetica. 1997. PMID: 9440270 Review.
-
Maintenance of transposable element copy number in natural populations of Drosophila melanogaster and D. simulans.Genetica. 1997;100(1-3):161-6. Genetica. 1997. PMID: 9440269 Review.
Cited by
-
Taming, Domestication and Exaptation: Trajectories of Transposable Elements in Genomes.Cells. 2021 Dec 20;10(12):3590. doi: 10.3390/cells10123590. Cells. 2021. PMID: 34944100 Free PMC article. Review.
-
The Interplay Between Developmental Stage and Environment Underlies the Adaptive Effect of a Natural Transposable Element Insertion.Mol Biol Evol. 2023 Mar 4;40(3):msad044. doi: 10.1093/molbev/msad044. Mol Biol Evol. 2023. PMID: 36811953 Free PMC article.
-
A benchmark of transposon insertion detection tools using real data.Mob DNA. 2019 Dec 30;10:53. doi: 10.1186/s13100-019-0197-9. eCollection 2019. Mob DNA. 2019. PMID: 31892957 Free PMC article.
References
-
- Modolo L, Lerat E. In: Identification and Analysis of transposable elements in genomic sequences. Poptsova MS, editor. U.K: Caister Academic Press; 2014.
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

