Regulatory Factors for tRNA Modifications in Extreme- Thermophilic Bacterium Thermus thermophilus
- PMID: 30906314
- PMCID: PMC6418473
- DOI: 10.3389/fgene.2019.00204
Regulatory Factors for tRNA Modifications in Extreme- Thermophilic Bacterium Thermus thermophilus
Abstract
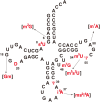
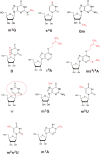
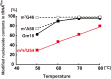
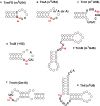
Thermus thermophilus is an extreme-thermophilic bacterium that can grow at a wide range of temperatures (50-83°C). To enable T. thermophilus to grow at high temperatures, several biomolecules including tRNA and tRNA modification enzymes show extreme heat-resistance. Therefore, the modified nucleosides in tRNA from T. thermophilus have been studied mainly from the view point of tRNA stabilization at high temperatures. Such studies have shown that several modifications stabilize the structure of tRNA and are essential for survival of the organism at high temperatures. Together with tRNA modification enzymes, the modified nucleosides form a network that regulates the extent of different tRNA modifications at various temperatures. In this review, I describe this network, as well as the tRNA recognition mechanism of individual tRNA modification enzymes. Furthermore, I summarize the roles of other tRNA stabilization factors such as polyamines and metal ions.
Keywords: RNA modification; Thermus thermophilus; methylation; tRNA; thermophile.
Figures

Similar articles
-
Effects of polyamines from Thermus thermophilus, an extreme-thermophilic eubacterium, on tRNA methylation by tRNA (Gm18) methyltransferase (TrmH).J Biochem. 2016 May;159(5):509-17. doi: 10.1093/jb/mvv130. Epub 2015 Dec 31. J Biochem. 2016. PMID: 26721905
-
Transfer RNA Modification Enzymes from Thermophiles and Their Modified Nucleosides in tRNA.Microorganisms. 2018 Oct 20;6(4):110. doi: 10.3390/microorganisms6040110. Microorganisms. 2018. PMID: 30347855 Free PMC article. Review.
-
Folate-/FAD-dependent tRNA methyltransferase from Thermus thermophilus regulates other modifications in tRNA at low temperatures.Genes Cells. 2016 Jul;21(7):740-54. doi: 10.1111/gtc.12376. Epub 2016 May 30. Genes Cells. 2016. PMID: 27238446
-
In vitro dihydrouridine formation by tRNA dihydrouridine synthase from Thermus thermophilus, an extreme-thermophilic eubacterium.J Biochem. 2015 Dec;158(6):513-21. doi: 10.1093/jb/mvv066. Epub 2015 Jun 24. J Biochem. 2015. PMID: 26112661
-
Dynamic structures and functions of transfer ribonucleic acids from extreme thermophiles.Adv Biophys. 1987;23:115-47. doi: 10.1016/0065-227x(87)90006-2. Adv Biophys. 1987. PMID: 2449804 Review.
Cited by
-
Functions of Bacterial tRNA Modifications: From Ubiquity to Diversity.Trends Microbiol. 2021 Jan;29(1):41-53. doi: 10.1016/j.tim.2020.06.010. Epub 2020 Jul 25. Trends Microbiol. 2021. PMID: 32718697 Free PMC article. Review.
-
Required Elements in tRNA for Methylation by the Eukaryotic tRNA (Guanine-N2-) Methyltransferase (Trm11-Trm112 Complex).Int J Mol Sci. 2022 Apr 6;23(7):4046. doi: 10.3390/ijms23074046. Int J Mol Sci. 2022. PMID: 35409407 Free PMC article.
-
Transfer RNA Modification Enzymes with a Thiouridine Synthetase, Methyltransferase and Pseudouridine Synthase (THUMP) Domain and the Nucleosides They Produce in tRNA.Genes (Basel). 2023 Jan 31;14(2):382. doi: 10.3390/genes14020382. Genes (Basel). 2023. PMID: 36833309 Free PMC article. Review.
-
The Thermus thermophilus DEAD-box protein Hera is a general RNA binding protein and plays a key role in tRNA metabolism.RNA. 2020 Nov;26(11):1557-1574. doi: 10.1261/rna.075580.120. Epub 2020 Jul 15. RNA. 2020. PMID: 32669294 Free PMC article.
-
Post-Transcriptional Modifications of Conserved Nucleotides in the T-Loop of tRNA: A Tale of Functional Convergent Evolution.Genes (Basel). 2021 Jan 22;12(2):140. doi: 10.3390/genes12020140. Genes (Basel). 2021. PMID: 33499018 Free PMC article. Review.
References
-
- Anantharaman V., Koonin E. V., Aravind L. (2002). SPOUT: a class of methyltransferases that includes spoU and trmD RNA methylase superfamilies, and novel superfamilies of predicted prokaryotic RNA methylases. J. Mol. Microbiol. Biotechnol. 4 71–75. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

