HPV type 16 E6 and NFX1-123 augment JNK signaling to mediate keratinocyte differentiation and L1 expression
- PMID: 30903928
- PMCID: PMC6486444
- DOI: 10.1016/j.virol.2019.03.008
HPV type 16 E6 and NFX1-123 augment JNK signaling to mediate keratinocyte differentiation and L1 expression
Abstract
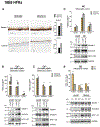
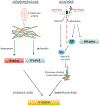
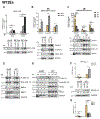
The HPV life cycle is differentiation-dependent, with cellular differentiation driving initiation of the late, productive stage of the viral life cycle. Here, we identify a role for the protein NFX1-123 in regulating keratinocyte differentiation and events of the late HPV life cycle. NFX1-123 itself increased with differentiation of epithelial cells. Greater NFX1-123 augmented differentiation marker expression and JNK phosphorylation in differentiating 16E6-expressing human foreskin keratinocytes (16E6 HFKs). This was associated with altered expression of MKK4 and MKK7, upstream kinase regulators of JNK phosphorylation. Modulating levels of NFX1-123 in HPV16-positive W12E cells recapitulated the effects on differentiation markers, JNK phosphorylation, and MKK4/7 seen in 16E6 HFKs. Crucially, levels of NFX1-123 also correlated with expression of L1, the capsid protein of HPV. Altogether, these studies define a role for NFX1-123 in mediating epithelial differentiation through the JNK signaling pathway, potentially linking expression of cellular genes and HPV genes during differentiation.
Keywords: Differentiation; HR E6; HR HPV; Keratinocyte; NFX1–123.
Copyright © 2019 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
NFX1, Its Isoforms and Roles in Biology, Disease and Cancer.Biology (Basel). 2021 Mar 30;10(4):279. doi: 10.3390/biology10040279. Biology (Basel). 2021. PMID: 33808060 Free PMC article. Review.
-
NFX1-123 and human papillomavirus 16E6 increase Notch expression in keratinocytes.J Virol. 2013 Dec;87(24):13741-50. doi: 10.1128/JVI.02582-13. Epub 2013 Oct 9. J Virol. 2013. PMID: 24109236 Free PMC article.
-
Cytoplasmic poly(A) binding proteins regulate telomerase activity and cell growth in human papillomavirus type 16 E6-expressing keratinocytes.J Virol. 2010 Dec;84(24):12934-44. doi: 10.1128/JVI.01377-10. Epub 2010 Oct 13. J Virol. 2010. PMID: 20943973 Free PMC article.
-
Human papillomavirus type 16 E6 and NFX1-123 mislocalize immune signaling proteins and downregulate immune gene expression in keratinocytes.PLoS One. 2017 Nov 8;12(11):e0187514. doi: 10.1371/journal.pone.0187514. eCollection 2017. PLoS One. 2017. PMID: 29117186 Free PMC article.
-
Post-Transcriptional Gene Regulation by HPV 16E6 and Its Host Protein Partners.Viruses. 2022 Jul 6;14(7):1483. doi: 10.3390/v14071483. Viruses. 2022. PMID: 35891463 Free PMC article. Review.
Cited by
-
NFX1-123: A potential therapeutic target in cervical cancer.J Med Virol. 2023 Jun;95(6):e28856. doi: 10.1002/jmv.28856. J Med Virol. 2023. PMID: 37288708 Free PMC article.
-
The complexity of human papilloma virus in cancers: a narrative review.Infect Agent Cancer. 2023 Feb 26;18(1):13. doi: 10.1186/s13027-023-00488-w. Infect Agent Cancer. 2023. PMID: 36843070 Free PMC article. Review.
-
High expression of NFX1-123 in HPV positive head and neck squamous cell carcinomas.Head Neck. 2022 Jan;44(1):177-188. doi: 10.1002/hed.26906. Epub 2021 Oct 25. Head Neck. 2022. PMID: 34693597 Free PMC article.
-
Genes Regulated by HPV 16 E6 and High Expression of NFX1-123 in Cervical Cancers.Onco Targets Ther. 2020 Jun 26;13:6143-6156. doi: 10.2147/OTT.S251926. eCollection 2020. Onco Targets Ther. 2020. PMID: 32617009 Free PMC article.
-
NFX1, Its Isoforms and Roles in Biology, Disease and Cancer.Biology (Basel). 2021 Mar 30;10(4):279. doi: 10.3390/biology10040279. Biology (Basel). 2021. PMID: 33808060 Free PMC article. Review.
References
-
- Doorbar J, Quint W, Banks L, Bravo IG, Stoler M, Broker TR, Stanley MA. 2012. The Biology and Life-Cycle of Human Papillomaviruses. Vaccine 30, Supplement 5:F55–F70. - PubMed
-
- zur Hausen H 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer 2:342–350. - PubMed
-
- Muñoz N, Bosch FX, de Sanjosé S, Herrero R, Castellsagué X, Shah KV, Snijders PJF, Meijer CJLM, International Agency for Research on Cancer Multicenter Cervical Cancer Study Group. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med 348:518–527. - PubMed
-
- zur Hausen H 2009. Papillomaviruses in the causation of human cancers — a brief historical account. Virology 384:260–265. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

