Autophagy-dependent secretion: mechanism, factors secreted, and disease implications
- PMID: 30894055
- PMCID: PMC6735501
- DOI: 10.1080/15548627.2019.1596479
Autophagy-dependent secretion: mechanism, factors secreted, and disease implications
Abstract
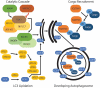
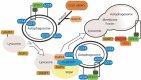
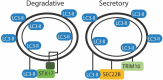
Although best understood as a degradative pathway, recent evidence demonstrates pronounced involvement of the macroautophagic/autophagic molecular machinery in cellular secretion. With either overexpression or inhibition of autophagy mediators, dramatic alterations in the cellular secretory profile occur. This affects secretion of a plethora of factors ranging from cytokines, to granule contents, and even viral particles. Encompassing a wide range of secreted factors, autophagy-dependent secretion is implicated in diseases ranging from cancer to neurodegeneration. With a growing body of evidence shedding light onto the molecular mediators, this review delineates the molecular machinery involved in selective targeting of the autophagosome for either degradation or secretion. In addition, we summarize the current understanding of factors and cargo secreted through this unconventional route, and describe the implications of this pathway in both health and disease. Abbreviations: BECN1, beclin 1; CAF, cancer associated fibroblast; CUPS, compartment for unconventional protein secretion; CXCL, C-X-C motif chemokine ligand; ER, endoplasmic reticulum; FGF2, fibroblast growth factor 2; HMGB1, high mobility group box 1; IDE, insulin degrading enzyme; IL, Interleukin; MAP1LC3/LC3, microtubule associated protein 1 light chain 3; MAPS, misfolding associated protein secretion; MEF, mouse embryonic fibroblast; MTORC1, MTOR complex I; PtdIns, phosphatidyl inositol; SEC22B, SEC22 homolog B, vesicle trafficking protein (gene/pseudogene); SFV, Semliki forest virus; SNCA, synuclein alpha; SQSTM1, sequestosome 1; STX, Syntaxin; TASCC, TOR-associated spatial coupling compartment; TGFB, transforming growth factor beta; TRIM16, tripartite motif containing 16; UPS, unconventional protein secretion; VWF, von Willebrand factor.
Keywords: Autophagy-dependent secretion; IL1B; cancer; infection; neurodegeneration; secretory autophagy.
Figures
Similar articles
-
Autophagy-based unconventional secretion of HMGB1 by keratinocytes plays a pivotal role in psoriatic skin inflammation.Autophagy. 2021 Feb;17(2):529-552. doi: 10.1080/15548627.2020.1725381. Epub 2020 Feb 16. Autophagy. 2021. PMID: 32019420 Free PMC article.
-
6-Hydroxydopamine induces secretion of PARK7/DJ-1 via autophagy-based unconventional secretory pathway.Autophagy. 2018;14(11):1943-1958. doi: 10.1080/15548627.2018.1493043. Epub 2018 Aug 16. Autophagy. 2018. PMID: 30112966 Free PMC article.
-
The GST-BHMT assay reveals a distinct mechanism underlying proteasome inhibition-induced macroautophagy in mammalian cells.Autophagy. 2015;11(5):812-32. doi: 10.1080/15548627.2015.1034402. Autophagy. 2015. PMID: 25984893 Free PMC article.
-
Impaired autophagy and APP processing in Alzheimer's disease: The potential role of Beclin 1 interactome.Prog Neurobiol. 2013 Jul-Aug;106-107:33-54. doi: 10.1016/j.pneurobio.2013.06.002. Epub 2013 Jul 1. Prog Neurobiol. 2013. PMID: 23827971 Review.
-
Secretory Autophagy and Its Relevance in Metabolic and Degenerative Disease.Front Endocrinol (Lausanne). 2020 May 5;11:266. doi: 10.3389/fendo.2020.00266. eCollection 2020. Front Endocrinol (Lausanne). 2020. PMID: 32477265 Free PMC article. Review.
Cited by
-
Migratory autolysosome disposal mitigates lysosome damage.J Cell Biol. 2024 Dec 2;223(12):e202403195. doi: 10.1083/jcb.202403195. Epub 2024 Sep 30. J Cell Biol. 2024. PMID: 39347717
-
Human platelets display dysregulated sepsis-associated autophagy, induced by altered LC3 protein-protein interaction of the Vici-protein EPG5.Autophagy. 2022 Jul;18(7):1534-1550. doi: 10.1080/15548627.2021.1990669. Epub 2021 Nov 18. Autophagy. 2022. PMID: 34689707 Free PMC article.
-
Lipopolysaccharide induces neuroinflammation in microglia by activating the MTOR pathway and downregulating Vps34 to inhibit autophagosome formation.J Neuroinflammation. 2020 Jan 11;17(1):18. doi: 10.1186/s12974-019-1644-8. J Neuroinflammation. 2020. PMID: 31926553 Free PMC article.
-
Autophagy-Based Unconventional Secretory for AIM2 Inflammasome Drives DNA Damage Resistance During Intervertebral Disc Degeneration.Front Cell Dev Biol. 2021 Jun 22;9:672847. doi: 10.3389/fcell.2021.672847. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34239872 Free PMC article.
-
The Role Played by Autophagy in FcεRI-Dependent Activation of Mast Cells.Cells. 2024 Apr 16;13(8):690. doi: 10.3390/cells13080690. Cells. 2024. PMID: 38667305 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
