Inhibition of proteasome rescues a pathogenic variant of respiratory chain assembly factor COA7
- PMID: 30885959
- PMCID: PMC6505684
- DOI: 10.15252/emmm.201809561
Inhibition of proteasome rescues a pathogenic variant of respiratory chain assembly factor COA7
Abstract
Nuclear and mitochondrial genome mutations lead to various mitochondrial diseases, many of which affect the mitochondrial respiratory chain. The proteome of the intermembrane space (IMS) of mitochondria consists of several important assembly factors that participate in the biogenesis of mitochondrial respiratory chain complexes. The present study comprehensively analyzed a recently identified IMS protein cytochrome c oxidase assembly factor 7 (COA7), or RESpiratory chain Assembly 1 (RESA1) factor that is associated with a rare form of mitochondrial leukoencephalopathy and complex IV deficiency. We found that COA7 requires the mitochondrial IMS import and assembly (MIA) pathway for efficient accumulation in the IMS We also found that pathogenic mutant versions of COA7 are imported slower than the wild-type protein, and mislocalized proteins are degraded in the cytosol by the proteasome. Interestingly, proteasome inhibition rescued both the mitochondrial localization of COA7 and complex IV activity in patient-derived fibroblasts. We propose proteasome inhibition as a novel therapeutic approach for a broad range of mitochondrial pathologies associated with the decreased levels of mitochondrial proteins.
Keywords: COA7/RESA1; mitochondrial disease; proteasome; protein degradation; protein import.
© 2019 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

- A
Protein abundance is the normalized signal intensity (LFQ) for a protein divided by its molecular weight. Specificity (enrichment) is the ratio of the protein LFQ intensity in the MIA40FLAG fraction to control samples. The LFQ for proteins that were not detected in the control samples was arbitrarily set to 1 for calculation purposes. LFQ, label‐free quantification; M.W., molecular weight.
- B
Flp‐In T‐REx 293 cells induced to express MIA40FLAG were solubilized, and the affinity purification of MIA40FLAG was performed. Fractions were analyzed by SDS–PAGE and Western blot. Load: 2.5%. Eluate: 100%. Unbound: 2.5%.
- C
Flp‐In T‐REx 293 cells induced to express MIA40FLAG were solubilized, and the affinity purification of MIA40FLAG was performed. Eluate fractions were solubilized under reducing (DTT) or non‐reducing (IAA) conditions and analyzed by SDS–PAGE and Western blot. DTT, dithiothreitol; IAA, iodoacetamide. Eluate: 100%.
- D
Cellular protein extracts from Flp‐In T‐REx 293 cells with induced expression of wild‐type or mutant MIA40FLAG were subjected to affinity purification. Load and eluate fractions were analyzed by reducing SDS–PAGE and Western blot. Load: 2.5%. Eluate: 100%.

- A
Flp‐In T‐REx 293 cells induced to express MIA40FLAG were solubilized, and the affinity purification of MIA40FLAG was performed. The fractions were analyzed by SDS–PAGE and Western blot. Load: 2.5%. Eluate: 100%.
- B
Flp‐In T‐REx 293 cells induced to express ALRFLAG were solubilized, and the affinity purification of ALRFLAG was performed. The fractions were analyzed by SDS–PAGE and Western blot. Load: 2.5%. Eluate: 100%.
- C
Mitochondria were isolated from Flp‐In T‐REx 293 cells induced to express wild‐type and mutant forms of MIA40FLAG under non‐reducing conditions. The extract was analyzed by non‐reducing SDS–PAGE and Western blot.
- D
Cellular protein extracts were isolated from Flp‐In T‐REx 293 cells induced to express wild‐type and mutant forms of MIA40FLAG under non‐reducing conditions. The extract was analyzed by non‐reducing SDS–PAGE and Western blot.
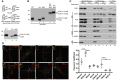
- A
Schematic representation of the thiol trapping assay. Mitochondria were solubilized in sample buffer with either dithiothreitol (DTT), iodoacetamide (IAA), or 4‐acetamido‐4‐maleimidylstilbene‐2,2‐disulfonic acid (AMS). The samples were analyzed by SDS–PAGE and Western blot.
- B
Indirect thiol trapping assay. Mitochondria were pretreated with IAA as indicated to block free cysteine residues, and disulfide bonds were subsequently reduced by DTT. Mitochondria were solubilized in sample buffer with AMS.
- C
Localization of mitochondrial proteins analyzed by limited degradation by proteinase K in intact mitochondria (250 mM sucrose), mitoplasts (5 mM sucrose), and mitochondrial lysates (1% Triton X‐100). The samples were analyzed by SDS–PAGE and Western blot. Mitos, mitochondria; Sup, post‐mitochondria supernatant; OM, outer membrane; IM, inner membrane; IMS, intermembrane space.
- D
N‐SIM super‐resolution micrographs of one Z‐stack (0.15 μm) orthogonal section (XYZ) of HeLa cells or HeLa cells that stably expressed COA7‐HA transfected with different markers TOMM20‐DsRed (OM), COX8A‐DsRed (IM), and matrix targeted photoactivatable GFP (Matrix target) labeled with anti‐HA, anti‐COA7, and Smac/Diablo (IMS) antibodies. The picture represents the majority population of cells from three independent experiments. Scale bar = 2 μm, scale bar in the magnified insert = 0.5 μm. The panel shows Pearson's coefficient in a co‐localized volume of different subcompartment combinations with anti‐HA or anti‐COA7. The data are expressed as a mean ± SD (n = 5). ***P < 0.001 [IMSxEnd vs. IMxEnd P = 0.0001; IMSxEnd vs. MxEnd P = 0.0002], ****P < 0.0001 [OMxHA vs. IMSxHA P < 0.0001; IMSxHA vs. IMxHA P < 0.0001] (one‐way ANOVA). M, matrix; End, endogenous COA7; HA, COA7‐HA.

- A
Mitochondria were isolated from cells that were transfected with a plasmid that encoded COA7HIS or an empty vector. Mitochondria were solubilized and analyzed by reducing SDS–PAGE and Western blot. Mitos, mitochondria.
- B
Radiolabeled [35S]TIMM8A and [35S]COX19 precursors were imported into mitochondria that were isolated from cells that were transfected with a plasmid that encoded COA7HIS or an empty vector. The samples were analyzed by reducing SDS–PAGE and autoradiography. The results of three biological replicates were analyzed, quantified, and normalized to control mitochondria at 30 min. The data are expressed as a mean ± SEM (n = 3). IAA, iodoacetamide.
- C
Mitochondria were isolated from cells that were transfected with a plasmid that encoded COA7HIS or an empty vector under reducing (DTT) and non‐reducing (IAA) conditions and analyzed for levels of MIA40 by Western blot.
- D
Proteins from HEK293 cells transfected with empty plasmid or COA7HIS were modified with PEG‐PCMal. Control cells were pretreated with the thiol‐oxidizing agent diamide (DAM) or the reductant DTT. The samples were analyzed by SDS–PAGE and Western blot.
- E
Flp‐In T‐REx 293 cells induced to express MIA40FLAG were transfected with a plasmid that encoded COA7HIS or an empty vector. The affinity purification of MIA40FLAG was performed, and eluate fractions were analyzed by SDS–PAGE and Western blot.
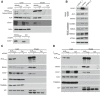
- A
Protein extracts were isolated from Flp‐In T‐REx 293 cells induced to express MIA40FLAG that was transfected with oligonucleotides that targeted COA7 mRNA or control oligonucleotides and subjected to affinity purification. The samples were analyzed by SDS–PAGE and Western blot. Load: 2.5%. Eluate: 100%.
- B
Cellular protein extracts were isolated from HeLa cells that were transfected with oligonucleotides that targeted different regions of COA7 mRNA or control oligonucleotides. The samples were analyzed by reducing SDS–PAGE and Western blot.
- C
Protein extracts were isolated from Flp‐In T‐REx 293 cells induced to express ALRFLAG that was transfected with oligonucleotides that targeted COA7 mRNA or control oligonucleotides and subjected to affinity purification. The samples were analyzed by SDS–PAGE and Western blot. Load: 2.5%. Eluate: 100%.
- D
Flp‐In T‐REx 293 cells induced to express ALRFLAG were transfected with a plasmid that encoded COA7HIS or an empty vector. The affinity purification of ALRFLAG was performed, and samples were analyzed by SDS–PAGE and Western blot.
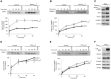
- A, B
Radiolabeled [35S]COA7 (A) and [35S]TIMM8A (B) precursors were imported into mitochondria that were isolated from Flp‐In T‐REx 293 cells induced to express MIA40FLAG. The samples were analyzed by reducing SDS–PAGE and autoradiography. The results of three biological replicates were analyzed, quantified, and normalized to control mitochondria at 45 min. The data are expressed as a mean ± SEM (n = 3). IAA, iodoacetamide.
- C
Mitochondria were isolated from Flp‐In T‐REx 293 cells induced to express MIA40FLAG and control cells. The samples were analyzed by SDS–PAGE and Western blot. Mitos, mitochondria.
- D, E
Radiolabeled [35S] COA7 (D) and [35S] TIMM8A (E) precursors were imported into mitochondria that were isolated from Flp‐In T‐REx 293 cells induced to express ALRFLAG and control cells. The samples were analyzed by reducing SDS–PAGE and autoradiography. The results of three biological experiments were analyzed, quantified, and normalized to control mitochondria at 45 min. The data are expressed as a mean ± SEM (n = 3).
- F
Mitochondria were isolated from Flp‐In T‐REx 293 cells induced to express ALRFLAG and control cells. The samples were analyzed by SDS–PAGE and Western blot.

- A
Cellular protein extracts were isolated from HeLa cells transfected with oligonucleotides that targeted different regions of MIA40 mRNA or with control oligonucleotides. The samples were analyzed by reducing SDS–PAGE and Western blot.
- B
Cellular protein extracts were isolated from HeLa cells transfected with oligonucleotides that targeted different regions of ALR mRNA or with control oligonucleotides. The samples were analyzed by reducing SDS–PAGE and Western blot.
- C
Schematic representation of MIA40 sequence containing the CPC motif in HEK293 MIA40 WT/WT and HEK293 MIA40 WT/Del53‐60 cells.
- D
Cellular protein extracts were isolated from HEK293 MIA40 WT/WT and WT/Del53‐60 cells. The samples were analyzed by SDS–PAGE and Western blot.
- E
Cellular protein extracts were isolated from HEK293 MIA40 WT/WT and WT/Del53‐60 cells transfected with a plasmid that encoded MIA40 or an empty vector. The samples were subjected to reducing SDS–PAGE and Western blot.
- F, G
Radiolabeled [35S]COA7 (F) and [35S]TIMM8A (G) precursors were imported into mitochondria that were isolated from HEK293 MIA40 WT/WT and WT/Del53‐60 cells. The samples were analyzed by reducing SDS–PAGE and autoradiography. The results of three biological replicates were analyzed, quantified, and normalized to control mitochondria at 45 min. The data are expressed as a mean ± SEM (n = 3). IAA, iodoacetamide.

- A
Schematic representation of wild‐type and mutant COA7.
- B
Cellular protein extracts were isolated from HEK293 cells that were transfected with a plasmid that encoded wild‐type or mutant COA7. The samples were analyzed by reducing SDS–PAGE and Western blot.
- C
Cellular fractions were prepared from HEK293 cells that were transfected with a plasmid that encoded wild‐type or mutant COA7. The fractions were analyzed by reducing SDS–PAGE and Western blot. T, total; C, cytosol; M, mitochondria.
- D
HEK293 cells that transiently expressed wild‐type or mutant COA7HIS were solubilized, and the affinity purification of COA7HIS was performed. The samples were analyzed by reducing and non‐reducing SDS–PAGE and Western blot. Load: 3%. Eluate: 100%.
- E
Equal amounts of wild‐type and mutant radiolabeled [35S]COA7 precursors were imported into mitochondria isolated from HEK293 cells. The samples were analyzed by reducing SDS–PAGE and autoradiography. The results of three biological replicates were analyzed, quantified, and normalized to wild‐type COA7 at 45 min. The data are expressed as a mean ± SEM (n = 3). IAA, iodoacetamide.

- A
Schematic representation of protein stability assay.
- B
HEK293 cells that transiently expressed wild‐type or mutant COA7HIS were treated with CHX for the indicated times, and protein extracts were isolated. The samples were analyzed by reducing SDS–PAGE and Western blot.
- C
Schematic representation of combined translation and proteasome inhibition assay.
- D
HEK293 cells that transiently expressed wild‐type or mutant COA7HIS were treated with CHX and/or MG132 for the indicated times, and protein extracts were isolated. The samples were analyzed by reducing SDS–PAGE and Western blot. CHX, cycloheximide.
- E
Schematic representation of the cellular localization assay.
- F
Cellular fractions were prepared from HEK293 cells that transiently expressed mutant COA7HIS and were treated with MG132. The samples were analyzed by reducing SDS–PAGE and Western blot.
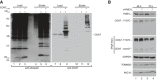
- A
Cellular protein extracts were isolated from HEK293 cells that expressed ubiquitinHIS (UbHis) and COA7FLAG and subjected to affinity purification. Load: 2.5%. Eluate: 100%.
- B
Cellular protein extracts were isolated after 48 and 72 h from patient fibroblast that were transfected with oligonucleotides that targeted different regions of YME1L mRNA or control oligonucleotides. The samples were analyzed by reducing SDS–PAGE and Western blot.

- A
Cellular protein extracts were isolated from immortalized patient‐derived skin fibroblasts that were treated with the indicated concentrations of MG132, bortezomib, or carfilzomib. The samples were analyzed by reducing SDS–PAGE and Western blot.
- B
Cellular fractions were prepared from immortalized patient‐derived skin fibroblasts that were treated with the indicated concentrations of MG132, bortezomib, or carfilzomib. The samples were analyzed by reducing SDS–PAGE and Western blot. T, total; C, cytosol; M, mitochondria.
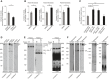
- A
Complex IV activity was assessed by oxidation of cytochrome c in digitonized cellular extracts from the immortalized patient‐derived skin fibroblasts and control healthy fibroblasts. The results of three biological replicates were quantified and normalized to control fibroblasts. Data are expressed as a mean ± SEM (n = 3, *P = 0.01) (two‐tailed Student's t‐test).
- B
Complex IV activity was assessed by oxidation of cytochrome c in digitonized cellular extracts from immortalized patient‐derived skin fibroblasts treated with MG132 (1 μM), bortezomib (20 nM), or carfilzomib (50 nM) for 12 h and recovered for another 6 h. The results of three biological replicates were quantified and normalized to DMSO‐treated samples, and the data are expressed as a mean ± SEM (n = 3, *P = 0.04 [DMSO vs MG132], **P = 0.0025 [DMSO vs Bortezomib], *P = 0.02 [DMSO vs Carfilzomib]) (two‐tailed Student's t‐test).
- C
Immortalized patient‐derived skin fibroblasts transiently expressing wild‐type or mutant COA7 (COA7‐Y137C and COA7‐Ex2∆) for 48 h were treated with bortezomib (20 nM) during the last 12 h. The fibroblasts were harvested, and complex IV activity was measured in digitonized cellular extracts. The results of three biological replicates were quantified and normalized to empty vector bortezomib‐treated samples, and the data are expressed as a mean ± SEM (n = 3, *P = 0.01 [Empty vector vs. COA7‐ Wt], *P = 0.005 [Empty vector vs COA7 – Y137C]) (two‐tailed Student's t‐test).
- D
Mitochondria isolated from control and patient fibroblast were solubilized in digitonin buffer and analyzed by 4–13% gel BN‐PAGE and Western blot. SC, supercomplexes.
- E
Immortalized patient‐derived skin fibroblasts transfected with plasmid encoding wild‐type COA7HIS or COA7‐Y137CHIS were solubilized in DDM buffer and analyzed by 4–13% gel BN‐PAGE and Western blot.
- F
Mitochondria were isolated from immortalized patient‐derived skin fibroblasts treated with bortezomib (10 nM) for 12 h and recovered for another 6 h. Mitochondria were solubilized in digitonin buffer and analyzed by 4–13% gel BN‐PAGE and Western blot. SC, supercomplexes.

- A
The precursor form of wild‐type COA7 is synthesized in the cytosol and imported into the mitochondrial intermembrane space by the MIA pathway, where it is involved in the respiratory chain biogenesis.
- B
The precursor form of mutant COA7 is import‐defective, and therefore, it is degraded in the cytosol by proteasome.
- C
Proteasome inhibition rescues the mitochondrial import of mutant COA7 as well as the biogenesis and function of the respiratory chain.
Similar articles
-
Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin.J Cell Biol. 2010 Dec 27;191(7):1367-80. doi: 10.1083/jcb.201007013. Epub 2010 Dec 20. J Cell Biol. 2010. PMID: 21173115 Free PMC article.
-
Hotspots for Disease-Causing Mutations in the Mitochondrial TIM23 Import Complex.Genes (Basel). 2024 Nov 28;15(12):1534. doi: 10.3390/genes15121534. Genes (Basel). 2024. PMID: 39766801 Free PMC article. Review.
-
Conserved quality control mechanisms of mitochondrial protein import.J Inherit Metab Dis. 2024 Sep;47(5):903-916. doi: 10.1002/jimd.12756. Epub 2024 May 24. J Inherit Metab Dis. 2024. PMID: 38790152 Review.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Adenosine Deaminase Deficiency.2006 Oct 3 [updated 2024 Mar 7]. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2025. 2006 Oct 3 [updated 2024 Mar 7]. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2025. PMID: 20301656 Free Books & Documents. Review.
Cited by
-
MitoStores: chaperone-controlled protein granules store mitochondrial precursors in the cytosol.EMBO J. 2023 Apr 3;42(7):e112309. doi: 10.15252/embj.2022112309. Epub 2023 Jan 27. EMBO J. 2023. PMID: 36704946 Free PMC article.
-
Proteolytic regulation of mitochondrial oxidative phosphorylation components in plants.Biochem Soc Trans. 2022 Jun 30;50(3):1119-1132. doi: 10.1042/BST20220195. Biochem Soc Trans. 2022. PMID: 35587610 Free PMC article. Review.
-
The intermembrane space protein Mix23 is a novel stress-induced mitochondrial import factor.J Biol Chem. 2020 Oct 23;295(43):14686-14697. doi: 10.1074/jbc.RA120.014247. Epub 2020 Aug 21. J Biol Chem. 2020. PMID: 32826315 Free PMC article.
-
The mitochondrial intermembrane space: the most constricted mitochondrial sub-compartment with the largest variety of protein import pathways.Open Biol. 2021 Mar;11(3):210002. doi: 10.1098/rsob.210002. Epub 2021 Mar 10. Open Biol. 2021. PMID: 33715390 Free PMC article. Review.
-
Proteasomal degradation induced by DPP9-mediated processing competes with mitochondrial protein import.EMBO J. 2020 Oct 1;39(19):e103889. doi: 10.15252/embj.2019103889. Epub 2020 Aug 20. EMBO J. 2020. PMID: 32815200 Free PMC article.
References
-
- Awasthi N, Wagner BJ (2005) Upregulation of heat shock protein expression by proteasome inhibition: an antiapoptotic mechanism in the lens. Invest Ophthalmol Vis Sci 46: 2082–2091 - PubMed
-
- Azad N, Vallyathan V, Wang L, Tantishaiyakul V, Stehlik C, Leonard SS, Rojanasakul Y (2006) S‐nitrosylation of Bcl‐2 inhibits its ubiquitin‐proteasomal degradation. A novel antiapoptotic mechanism that suppresses apoptosis. J Biol Chem 281: 34124–34134 - PubMed
-
- Banci L, Bertini I, Cefaro C, Ciofi‐Baffoni S, Gallo A, Martinelli M, Sideris DP, Katrakili N, Tokatlidis K (2009) MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria. Nat Struct Mol Biol 16: 198–206 - PubMed
-
- Banci L, Bertini I, Cefaro C, Cenacchi L, Ciofi‐Baffoni S, Felli IC, Gallo A, Gonnelli L, Luchinat E, Sideris D et al (2010) Molecular chaperone function of Mia40 triggers consecutive induced folding steps of the substrate in mitochondrial protein import. Proc Natl Acad Sci USA 107: 20190–20195 - PMC - PubMed
-
- Banci L, Bertini I, Calderone V, Cefaro C, Ciofi‐Baffoni S, Gallo A, Tokatlidis K (2012) An electron‐transfer path through an extended disulfide relay system: the case of the redox protein ALR. J Am Chem Soc 134: 1442–1445 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

