Cardiomyogenic differentiation is fine-tuned by differential mRNA association with polysomes
- PMID: 30876407
- PMCID: PMC6420765
- DOI: 10.1186/s12864-019-5550-3
Cardiomyogenic differentiation is fine-tuned by differential mRNA association with polysomes
Abstract
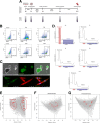
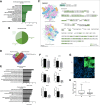
Background: Cardiac cell fate specification occurs through progressive steps, and its gene expression regulation features are still being defined. There has been an increasing interest in understanding the coordination between transcription and post-transcriptional regulation during the differentiation processes. Here, we took advantage of the polysome profiling technique to isolate and high-throughput sequence ribosome-free and polysome-bound RNAs during cardiomyogenesis.
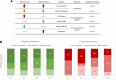
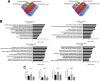
Results: We showed that polysome-bound RNAs exhibit the cardiomyogenic commitment gene expression and that mesoderm-to-cardiac progenitor stages are strongly regulated. Additionally, we compared ribosome-free and polysome-bound RNAs and found that the post-transcriptional regulation vastly contributes to cardiac phenotype determination, including RNA recruitment to and dissociation from ribosomes. Moreover, we found that protein synthesis is decreased in cardiomyocytes compared to human embryonic stem-cells (hESCs), possibly due to the down-regulation of translation-related genes.
Conclusions: Our data provided a powerful tool to investigate genes potentially controlled by post-transcriptional mechanisms during the cardiac differentiation of hESC. This work could prospect fundamental tools to develop new therapy and research approaches.
Keywords: Cardiomyogenesis; Polysome profiling; Post-transcriptional regulation; hESC-derived cardiomyocytes.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



Similar articles
-
Polysome profiling followed by RNA-seq of cardiac differentiation stages in hESCs.Sci Data. 2018 Dec 4;5:180287. doi: 10.1038/sdata.2018.287. Sci Data. 2018. PMID: 30512016 Free PMC article.
-
Polysome-associated lncRNAs during cardiomyogenesis of hESCs.Mol Cell Biochem. 2020 May;468(1-2):35-45. doi: 10.1007/s11010-020-03709-7. Epub 2020 Mar 3. Mol Cell Biochem. 2020. PMID: 32125578
-
Reorganization of Metabolism during Cardiomyogenesis Implies Time-Specific Signaling Pathway Regulation.Int J Mol Sci. 2021 Jan 29;22(3):1330. doi: 10.3390/ijms22031330. Int J Mol Sci. 2021. PMID: 33572750 Free PMC article.
-
Genome-Wide Transcriptome and Binding Sites Analyses Identify Early FOX Expressions for Enhancing Cardiomyogenesis Efficiency of hESC Cultures.Sci Rep. 2016 Aug 9;6:31068. doi: 10.1038/srep31068. Sci Rep. 2016. PMID: 27501774 Free PMC article.
-
Translation Analysis at the Genome Scale by Ribosome Profiling.Methods Mol Biol. 2016;1361:105-24. doi: 10.1007/978-1-4939-3079-1_7. Methods Mol Biol. 2016. PMID: 26483019 Review.
Cited by
-
Effects of PUMILIO1 and PUMILIO2 knockdown on cardiomyogenic differentiation of human embryonic stem cells culture.PLoS One. 2020 May 21;15(5):e0222373. doi: 10.1371/journal.pone.0222373. eCollection 2020. PLoS One. 2020. PMID: 32437472 Free PMC article.
-
Cardiomyogenesis Modeling Using Pluripotent Stem Cells: The Role of Microenvironmental Signaling.Front Cell Dev Biol. 2019 Aug 9;7:164. doi: 10.3389/fcell.2019.00164. eCollection 2019. Front Cell Dev Biol. 2019. PMID: 31448277 Free PMC article. Review.
-
Translational control of stem cell function.Nat Rev Mol Cell Biol. 2021 Oct;22(10):671-690. doi: 10.1038/s41580-021-00386-2. Epub 2021 Jul 16. Nat Rev Mol Cell Biol. 2021. PMID: 34272502 Review.
-
Quantitative proteomic profiling identifies global protein network dynamics in murine embryonic heart development.Dev Cell. 2023 Jun 19;58(12):1087-1105.e4. doi: 10.1016/j.devcel.2023.04.011. Epub 2023 May 5. Dev Cell. 2023. PMID: 37148880 Free PMC article.
-
Ribosome heterogeneity in stem cells and development.J Cell Biol. 2020 Jun 1;219(6):e202001108. doi: 10.1083/jcb.202001108. J Cell Biol. 2020. PMID: 32330234 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

