S-Geranylgeranyl-L-glutathione is a ligand for human B cell-confinement receptor P2RY8
- PMID: 30842656
- PMCID: PMC6640153
- DOI: 10.1038/s41586-019-1003-z
S-Geranylgeranyl-L-glutathione is a ligand for human B cell-confinement receptor P2RY8
Abstract
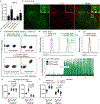
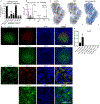
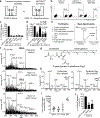
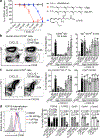
Germinal centres are important sites for antibody diversification and affinity maturation, and are also a common origin of B cell malignancies. Despite being made up of motile cells, germinal centres are tightly confined within B cell follicles. The cues that promote this confinement are incompletely understood. P2RY8 is a Gα13-coupled receptor that mediates the inhibition of migration and regulates the growth of B cells in lymphoid tissues1,2. P2RY8 is frequently mutated in germinal-centre B cell-like diffuse large B cell lymphoma (GCB-DLBCL) and Burkitt lymphoma1,3-6, and the ligand for this receptor has not yet been identified. Here we perform a search for P2RY8 ligands and find P2RY8 bioactivity in bile and in culture supernatants of several mouse and human cell lines. Using a seven-step biochemical fractionation procedure and a drop-out mass spectrometry approach, we show that a previously undescribed biomolecule, S-geranylgeranyl-L-glutathione (GGG), is a potent P2RY8 ligand that is detectable in lymphoid tissues at the nanomolar level. GGG inhibited the chemokine-mediated migration of human germinal-centre B cells and T follicular helper cells, and antagonized the induction of phosphorylated AKT in germinal-centre B cells. We also found that the enzyme gamma-glutamyltransferase-5 (GGT5), which was highly expressed by follicular dendritic cells, metabolized GGG to a form that did not activate the receptor. Overexpression of GGT5 disrupted the ability of P2RY8 to promote B cell confinement to germinal centres, which indicates that GGT5 establishes a GGG gradient in lymphoid tissues. This work defines GGG as an intercellular signalling molecule that is involved in organizing and controlling germinal-centre responses. As the P2RY8 locus is modified in several other types of cancer in addition to GCB-DLBCL and Burkitt lymphoma, we speculate that GGG might have organizing and growth-regulatory roles in multiple human tissues.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures







Comment in
-
Chemical choreography of germinal center B-cell migration.Cell Res. 2019 Jul;29(7):514-515. doi: 10.1038/s41422-019-0170-7. Cell Res. 2019. PMID: 31024167 Free PMC article. No abstract available.
Similar articles
-
Loss of signalling via Gα13 in germinal centre B-cell-derived lymphoma.Nature. 2014 Dec 11;516(7530):254-8. doi: 10.1038/nature13765. Epub 2014 Sep 28. Nature. 2014. PMID: 25274307 Free PMC article.
-
Abcc1 and Ggt5 support lymphocyte guidance through export and catabolism of S-geranylgeranyl-l-glutathione.Sci Immunol. 2021 Jun 4;6(60):eabg1101. doi: 10.1126/sciimmunol.abg1101. Sci Immunol. 2021. PMID: 34088745 Free PMC article.
-
The G protein-coupled receptor P2RY8 and follicular dendritic cells promote germinal center confinement of B cells, whereas S1PR3 can contribute to their dissemination.J Exp Med. 2015 Dec 14;212(13):2213-22. doi: 10.1084/jem.20151250. Epub 2015 Nov 16. J Exp Med. 2015. PMID: 26573295 Free PMC article.
-
G-protein coupled receptors and ligands that organize humoral immune responses.Immunol Rev. 2019 May;289(1):158-172. doi: 10.1111/imr.12743. Immunol Rev. 2019. PMID: 30977196 Free PMC article. Review.
-
Human T follicular helper cells: development and subsets.Adv Exp Med Biol. 2013;785:87-94. doi: 10.1007/978-1-4614-6217-0_10. Adv Exp Med Biol. 2013. PMID: 23456841 Review.
Cited by
-
Endoplasmic reticulum stress and glutathione therapeutics in chronic lung diseases.Redox Biol. 2020 Jun;33:101516. doi: 10.1016/j.redox.2020.101516. Epub 2020 Mar 23. Redox Biol. 2020. PMID: 32249209 Free PMC article. Review. No abstract available.
-
Glutathione-Dependent Pathways in Cancer Cells.Int J Mol Sci. 2024 Aug 1;25(15):8423. doi: 10.3390/ijms25158423. Int J Mol Sci. 2024. PMID: 39125992 Free PMC article. Review.
-
GPR35 promotes neutrophil recruitment in response to serotonin metabolite 5-HIAA.Cell. 2022 Mar 3;185(5):815-830.e19. doi: 10.1016/j.cell.2022.01.010. Epub 2022 Feb 10. Cell. 2022. PMID: 35148838 Free PMC article.
-
Integrative transcriptomic, proteomic, and machine learning approach to identifying feature genes of atrial fibrillation using atrial samples from patients with valvular heart disease.BMC Cardiovasc Disord. 2021 Jan 28;21(1):52. doi: 10.1186/s12872-020-01819-0. BMC Cardiovasc Disord. 2021. PMID: 33509101 Free PMC article.
-
P2RY8 variants in lupus patients uncover a role for the receptor in immunological tolerance.J Exp Med. 2022 Jan 3;219(1):e20211004. doi: 10.1084/jem.20211004. Epub 2021 Dec 10. J Exp Med. 2022. PMID: 34889940 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

