ABHD5 blunts the sensitivity of colorectal cancer to fluorouracil via promoting autophagic uracil yield
- PMID: 30842415
- PMCID: PMC6403256
- DOI: 10.1038/s41467-019-08902-x
ABHD5 blunts the sensitivity of colorectal cancer to fluorouracil via promoting autophagic uracil yield
Abstract
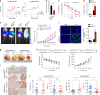
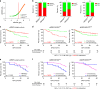
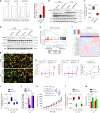
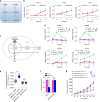
The efficacy of Fluorouracil (FU) in the treatment of colorectal cancer (CRC) is greatly limited by drug resistance. Autophagy has been implicated in chemoresistance, but the role of selective autophagic degradation in regulating chemoresistance remains unknown. In this study, we revealed a critical role of ABHD5 in charging CRC sensitivity to FU via regulating autophagic uracil yield. We demonstrated that ABHD5 localizes to lysosome and interacts with PDIA5 to prevent PDIA5 from interacting with RNASET2 and inactivating RNASET2. ABHD5 deficiency releases PDIA5 to directly interact with RNASET2 and leave RNASET2 in an inactivate state, which impairs RNASET2-mediated autophagic uracil yield and promotes CRC cells to uptake FU as an exogenous uracil, thus increasing their sensitivity to FU. Our findings for the first time reveal a novel role of ABHD5 in regulating lysosome function, highlighting the significance of ABHD5 as a compelling biomarker predicting the sensitivity of CRCs to FU-based chemotherapy.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Increased S1P induces S1PR2 internalization to blunt the sensitivity of colorectal cancer to 5-fluorouracil via promoting intracellular uracil generation.Acta Pharmacol Sin. 2021 Mar;42(3):460-469. doi: 10.1038/s41401-020-0460-0. Epub 2020 Jul 9. Acta Pharmacol Sin. 2021. PMID: 32647340 Free PMC article.
-
ABHD5 inhibits YAP-induced c-Met overexpression and colon cancer cell stemness via suppressing YAP methylation.Nat Commun. 2021 Nov 18;12(1):6711. doi: 10.1038/s41467-021-26967-5. Nat Commun. 2021. PMID: 34795238 Free PMC article.
-
The loss of SHMT2 mediates 5-fluorouracil chemoresistance in colorectal cancer by upregulating autophagy.Oncogene. 2021 Jun;40(23):3974-3988. doi: 10.1038/s41388-021-01815-4. Epub 2021 May 14. Oncogene. 2021. PMID: 33990700 Free PMC article.
-
ABHD5 interacts with BECN1 to regulate autophagy and tumorigenesis of colon cancer independent of PNPLA2.Autophagy. 2016 Nov;12(11):2167-2182. doi: 10.1080/15548627.2016.1217380. Epub 2016 Aug 25. Autophagy. 2016. PMID: 27559856 Free PMC article.
-
5-Fluorouracil targets histone acetyltransferases p300/CBP in the treatment of colorectal cancer.Cancer Lett. 2017 Aug 1;400:183-193. doi: 10.1016/j.canlet.2017.04.033. Epub 2017 Apr 29. Cancer Lett. 2017. PMID: 28465257
Cited by
-
Downregulation of Ascl2 promotes cell apoptosis by enhancing autophagy in colorectal cancer cells.J Gastrointest Oncol. 2021 Apr;12(2):630-638. doi: 10.21037/jgo-21-183. J Gastrointest Oncol. 2021. PMID: 34012655 Free PMC article.
-
ABHD5 as a friend or an enemy in cancer biology?Front Oncol. 2024 Sep 12;14:1447509. doi: 10.3389/fonc.2024.1447509. eCollection 2024. Front Oncol. 2024. PMID: 39328203 Free PMC article. Review.
-
From Intestinal Epithelial Homeostasis to Colorectal Cancer: Autophagy Regulation in Cellular Stress.Antioxidants (Basel). 2022 Jun 30;11(7):1308. doi: 10.3390/antiox11071308. Antioxidants (Basel). 2022. PMID: 35883800 Free PMC article. Review.
-
A Comprehensive Study on Folate-Targeted Mesoporous Silica Nanoparticles Loaded with 5-Fluorouracil for the Enhanced Treatment of Gynecological Cancers.J Funct Biomater. 2024 Mar 20;15(3):74. doi: 10.3390/jfb15030074. J Funct Biomater. 2024. PMID: 38535267 Free PMC article.
-
RUNX1-PDIA5 Axis Promotes Malignant Progression of Glioblastoma by Regulating CCAR1 Protein Expression.Int J Biol Sci. 2024 Aug 12;20(11):4364-4381. doi: 10.7150/ijbs.92595. eCollection 2024. Int J Biol Sci. 2024. PMID: 39247813 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

