Virus Genomes from Deep Sea Sediments Expand the Ocean Megavirome and Support Independent Origins of Viral Gigantism
- PMID: 30837339
- PMCID: PMC6401483
- DOI: 10.1128/mBio.02497-18
Virus Genomes from Deep Sea Sediments Expand the Ocean Megavirome and Support Independent Origins of Viral Gigantism
Abstract
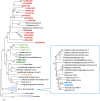
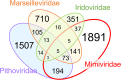
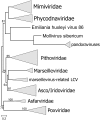
The nucleocytoplasmic large DNA viruses (NCLDV) of eukaryotes (proposed order, "Megavirales") include the families Poxviridae, Asfarviridae, Iridoviridae, Ascoviridae, Phycodnaviridae, Marseilleviridae, and Mimiviridae, as well as still unclassified pithoviruses, pandoraviruses, molliviruses, and faustoviruses. Several of these virus groups include giant viruses, with genome and particle sizes exceeding those of many bacterial and archaeal cells. We explored the diversity of the NCLDV in deep sea sediments from the Loki's Castle hydrothermal vent area. Using metagenomics, we reconstructed 23 high-quality genomic bins of novel NCLDV, 15 of which are related to pithoviruses, 5 to marseilleviruses, 1 to iridoviruses, and 2 to klosneuviruses. Some of the identified pithovirus-like and marseillevirus-like genomes belong to deep branches in the phylogenetic tree of core NCLDV genes, substantially expanding the diversity and phylogenetic depth of the respective groups. The discovered viruses, including putative giant members of the family Marseilleviridae, have a broad range of apparent genome sizes, in agreement with the multiple, independent origins of gigantism in different branches of the NCLDV. Phylogenomic analysis reaffirms the monophyly of the pithovirus-iridovirus-marseillevirus branch of the NCLDV. Similarly to other giant viruses, the pithovirus-like viruses from Loki's Castle encode translation systems components. Phylogenetic analysis of these genes indicates a greater bacterial contribution than had been detected previously. Genome comparison suggests extensive gene exchange between members of the pithovirus-like viruses and Mimiviridae Further exploration of the genomic diversity of Megavirales in additional sediment samples is expected to yield new insights into the evolution of giant viruses and the composition of the ocean megavirome.IMPORTANCE Genomics and evolution of giant viruses are two of the most vigorously developing areas of virus research. Lately, metagenomics has become the main source of new virus genomes. Here we describe a metagenomic analysis of the genomes of large and giant viruses from deep sea sediments. The assembled new virus genomes substantially expand the known diversity of the nucleocytoplasmic large DNA viruses of eukaryotes. The results support the concept of independent evolution of giant viruses from smaller ancestors in different virus branches.
Keywords: deep sea sediments; giant viruses; metagenomics; nucleocytoplasmic large DNA viruses; virus evolution.
Copyright © 2019 Bäckström et al.
Figures




Similar articles
-
Evolution of the Large Nucleocytoplasmic DNA Viruses of Eukaryotes and Convergent Origins of Viral Gigantism.Adv Virus Res. 2019;103:167-202. doi: 10.1016/bs.aivir.2018.09.002. Epub 2018 Nov 10. Adv Virus Res. 2019. PMID: 30635076 Review.
-
Origin of giant viruses from smaller DNA viruses not from a fourth domain of cellular life.Virology. 2014 Oct;466-467:38-52. doi: 10.1016/j.virol.2014.06.032. Epub 2014 Jul 17. Virology. 2014. PMID: 25042053 Free PMC article.
-
Characterization of Mollivirus kamchatka, the First Modern Representative of the Proposed Molliviridae Family of Giant Viruses.J Virol. 2020 Mar 31;94(8):e01997-19. doi: 10.1128/JVI.01997-19. Print 2020 Mar 31. J Virol. 2020. PMID: 31996429 Free PMC article.
-
Evidence of the megavirome in humans.J Clin Virol. 2013 Jul;57(3):191-200. doi: 10.1016/j.jcv.2013.03.018. Epub 2013 May 8. J Clin Virol. 2013. PMID: 23664726
-
Updating strategies for isolating and discovering giant viruses.Curr Opin Microbiol. 2016 Jun;31:80-87. doi: 10.1016/j.mib.2016.03.004. Epub 2016 Apr 1. Curr Opin Microbiol. 2016. PMID: 27039269 Review.
Cited by
-
Viral Diversity in Benthic Abyssal Ecosystems: Ecological and Methodological Considerations.Viruses. 2023 Nov 21;15(12):2282. doi: 10.3390/v15122282. Viruses. 2023. PMID: 38140524 Free PMC article.
-
Giant Viruses-Big Surprises.Viruses. 2019 Apr 30;11(5):404. doi: 10.3390/v11050404. Viruses. 2019. PMID: 31052218 Free PMC article. Review.
-
Giant virus biology and diversity in the era of genome-resolved metagenomics.Nat Rev Microbiol. 2022 Dec;20(12):721-736. doi: 10.1038/s41579-022-00754-5. Epub 2022 Jul 28. Nat Rev Microbiol. 2022. PMID: 35902763 Review.
-
Viral histones: pickpocket's prize or primordial progenitor?Epigenetics Chromatin. 2022 May 28;15(1):21. doi: 10.1186/s13072-022-00454-7. Epigenetics Chromatin. 2022. PMID: 35624484 Free PMC article. Review.
-
Expanding standards in viromics: in silico evaluation of dsDNA viral genome identification, classification, and auxiliary metabolic gene curation.PeerJ. 2021 Jun 14;9:e11447. doi: 10.7717/peerj.11447. eCollection 2021. PeerJ. 2021. PMID: 34178438 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

