Wide diversity of methane and short-chain alkane metabolisms in uncultured archaea
- PMID: 30833729
- PMCID: PMC6453112
- DOI: 10.1038/s41564-019-0363-3
Wide diversity of methane and short-chain alkane metabolisms in uncultured archaea
Abstract
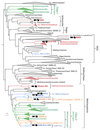
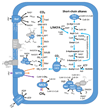
Methanogenesis is an ancient metabolism of key ecological relevance, with direct impact on the evolution of Earth's climate. Recent results suggest that the diversity of methane metabolisms and their derivations have probably been vastly underestimated. Here, by probing thousands of publicly available metagenomes for homologues of methyl-coenzyme M reductase complex (MCR), we have obtained ten metagenome-assembled genomes (MAGs) belonging to potential methanogenic, anaerobic methanotrophic and short-chain alkane-oxidizing archaea. Five of these MAGs represent under-sampled (Verstraetearchaeota, Methanonatronarchaeia, ANME-1 and GoM-Arc1) or previously genomically undescribed (ANME-2c) archaeal lineages. The remaining five MAGs correspond to lineages that are only distantly related to previously known methanogens and span the entire archaeal phylogeny. Comprehensive comparative annotation substantially expands the metabolic diversity and energy conservation systems of MCR-bearing archaea. It also suggests the potential existence of a yet uncharacterized type of methanogenesis linked to short-chain alkane/fatty acid oxidation in a previously undescribed class of archaea ('Candidatus Methanoliparia'). We redefine a common core of marker genes specific to methanogenic, anaerobic methanotrophic and short-chain alkane-oxidizing archaea, and propose a possible scenario for the evolutionary and functional transitions that led to the emergence of such metabolic diversity.
Conflict of interest statement
The authors declare no competing interests.
Figures


Comment in
-
Sediment, methane and energy.Nat Microbiol. 2019 Apr;4(4):547-549. doi: 10.1038/s41564-019-0417-6. Nat Microbiol. 2019. PMID: 30899110 No abstract available.
Similar articles
-
Anaerobic Degradation of Non-Methane Alkanes by "Candidatus Methanoliparia" in Hydrocarbon Seeps of the Gulf of Mexico.mBio. 2019 Aug 20;10(4):e01814-19. doi: 10.1128/mBio.01814-19. mBio. 2019. PMID: 31431553 Free PMC article.
-
"Candidatus Ethanoperedens," a Thermophilic Genus of Archaea Mediating the Anaerobic Oxidation of Ethane.mBio. 2020 Apr 21;11(2):e00600-20. doi: 10.1128/mBio.00600-20. mBio. 2020. PMID: 32317322 Free PMC article.
-
Expanding anaerobic alkane metabolism in the domain of Archaea.Nat Microbiol. 2019 Apr;4(4):595-602. doi: 10.1038/s41564-019-0364-2. Epub 2019 Mar 4. Nat Microbiol. 2019. PMID: 30833728 Review.
-
Diverse anaerobic methane- and multi-carbon alkane-metabolizing archaea coexist and show activity in Guaymas Basin hydrothermal sediment.Environ Microbiol. 2019 Apr;21(4):1344-1355. doi: 10.1111/1462-2920.14568. Epub 2019 Mar 18. Environ Microbiol. 2019. PMID: 30790413
-
Methyl/alkyl-coenzyme M reductase-based anaerobic alkane oxidation in archaea.Environ Microbiol. 2021 Feb;23(2):530-541. doi: 10.1111/1462-2920.15057. Epub 2020 May 18. Environ Microbiol. 2021. PMID: 32367670 Review.
Cited by
-
Genomic and transcriptomic insights into methanogenesis potential of novel methanogens from mangrove sediments.Microbiome. 2020 Jun 17;8(1):94. doi: 10.1186/s40168-020-00876-z. Microbiome. 2020. PMID: 32552798 Free PMC article.
-
TreeSAPP: the Tree-based Sensitive and Accurate Phylogenetic Profiler.Bioinformatics. 2020 Sep 15;36(18):4706-4713. doi: 10.1093/bioinformatics/btaa588. Bioinformatics. 2020. PMID: 32637989 Free PMC article.
-
Covariation of hot spring geochemistry with microbial genomic diversity, function, and evolution.Nat Commun. 2024 Aug 29;15(1):7506. doi: 10.1038/s41467-024-51841-5. Nat Commun. 2024. PMID: 39209850 Free PMC article.
-
Microbial Hydrocarbon Degradation in Guaymas Basin-Exploring the Roles and Potential Interactions of Fungi and Sulfate-Reducing Bacteria.Front Microbiol. 2022 Mar 9;13:831828. doi: 10.3389/fmicb.2022.831828. eCollection 2022. Front Microbiol. 2022. PMID: 35356530 Free PMC article.
-
Non-syntrophic methanogenic hydrocarbon degradation by an archaeal species.Nature. 2022 Jan;601(7892):257-262. doi: 10.1038/s41586-021-04235-2. Epub 2021 Dec 22. Nature. 2022. PMID: 34937940
References
-
- Demirel B, Scherer P. The roles of acetotrophic and hydrogenotrophic methanogens during anaerobic conversion of biomass to methane: A review. Rev Environ Sci Biotechnol. 2008;7:173–190.
-
- Ueno Y, Yamada K, Yoshida N, Maruyama S, Isozaki Y. Evidence from fluid inclusions for microbial methanogenesis in the early Archaean era. Nature. 2006;440:516–519. - PubMed
-
- Kasting JF, Siefert JL. Life and the evolution of Earth’s atmosphere. Science. 2002;296:1066–1068. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

