Stress response, behavior, and development are shaped by transposable element-induced mutations in Drosophila
- PMID: 30753202
- PMCID: PMC6372155
- DOI: 10.1371/journal.pgen.1007900
Stress response, behavior, and development are shaped by transposable element-induced mutations in Drosophila
Abstract
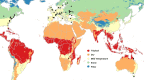
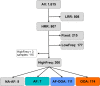
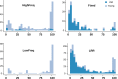
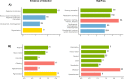
Most of the current knowledge on the genetic basis of adaptive evolution is based on the analysis of single nucleotide polymorphisms (SNPs). Despite increasing evidence for their causal role, the contribution of structural variants to adaptive evolution remains largely unexplored. In this work, we analyzed the population frequencies of 1,615 Transposable Element (TE) insertions annotated in the reference genome of Drosophila melanogaster, in 91 samples from 60 worldwide natural populations. We identified a set of 300 polymorphic TEs that are present at high population frequencies, and located in genomic regions with high recombination rate, where the efficiency of natural selection is high. The age and the length of these 300 TEs are consistent with relatively young and long insertions reaching high frequencies due to the action of positive selection. Besides, we identified a set of 21 fixed TEs also likely to be adaptive. Indeed, we, and others, found evidence of selection for 84 of these reference TE insertions. The analysis of the genes located nearby these 84 candidate adaptive insertions suggested that the functional response to selection is related with the GO categories of response to stimulus, behavior, and development. We further showed that a subset of the candidate adaptive TEs affects expression of nearby genes, and five of them have already been linked to an ecologically relevant phenotypic effect. Our results provide a more complete understanding of the genetic variation and the fitness-related traits relevant for adaptive evolution. Similar studies should help uncover the importance of TE-induced adaptive mutations in other species as well.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Sequencing of pooled DNA samples (Pool-Seq) uncovers complex dynamics of transposable element insertions in Drosophila melanogaster.PLoS Genet. 2012 Jan;8(1):e1002487. doi: 10.1371/journal.pgen.1002487. Epub 2012 Jan 26. PLoS Genet. 2012. PMID: 22291611 Free PMC article.
-
Population-specific dynamics and selection patterns of transposable element insertions in European natural populations.Mol Ecol. 2019 Mar;28(6):1506-1522. doi: 10.1111/mec.14963. Epub 2019 Jan 17. Mol Ecol. 2019. PMID: 30506554 Free PMC article.
-
High rate of recent transposable element-induced adaptation in Drosophila melanogaster.PLoS Biol. 2008 Oct 21;6(10):e251. doi: 10.1371/journal.pbio.0060251. PLoS Biol. 2008. PMID: 18942889 Free PMC article.
-
Population genomics of transposable elements in Drosophila.Annu Rev Genet. 2014;48:561-81. doi: 10.1146/annurev-genet-120213-092359. Epub 2014 Oct 1. Annu Rev Genet. 2014. PMID: 25292358 Review.
-
Transposable elements in natural populations of Drosophila melanogaster.Philos Trans R Soc Lond B Biol Sci. 2010 Apr 27;365(1544):1219-28. doi: 10.1098/rstb.2009.0318. Philos Trans R Soc Lond B Biol Sci. 2010. PMID: 20308097 Free PMC article. Review.
Cited by
-
ChimeraTE: a pipeline to detect chimeric transcripts derived from genes and transposable elements.Nucleic Acids Res. 2023 Oct 13;51(18):9764-9784. doi: 10.1093/nar/gkad671. Nucleic Acids Res. 2023. PMID: 37615575 Free PMC article.
-
The Structural, Functional and Evolutionary Impact of Transposable Elements in Eukaryotes.Genes (Basel). 2021 Jun 15;12(6):918. doi: 10.3390/genes12060918. Genes (Basel). 2021. PMID: 34203645 Free PMC article. Review.
-
Genomics of Evolutionary Novelty in Hybrids and Polyploids.Front Genet. 2020 Jul 28;11:792. doi: 10.3389/fgene.2020.00792. eCollection 2020. Front Genet. 2020. PMID: 32849797 Free PMC article. Review.
-
Measuring and interpreting transposable element expression.Nat Rev Genet. 2020 Dec;21(12):721-736. doi: 10.1038/s41576-020-0251-y. Epub 2020 Jun 23. Nat Rev Genet. 2020. PMID: 32576954 Review.
-
Population-scale long-read sequencing uncovers transposable elements associated with gene expression variation and adaptive signatures in Drosophila.Nat Commun. 2022 Apr 12;13(1):1948. doi: 10.1038/s41467-022-29518-8. Nat Commun. 2022. PMID: 35413957 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

