Labeling Strategies Matter for Super-Resolution Microscopy: A Comparison between HaloTags and SNAP-tags
- PMID: 30745239
- PMCID: PMC6474801
- DOI: 10.1016/j.chembiol.2019.01.003
Labeling Strategies Matter for Super-Resolution Microscopy: A Comparison between HaloTags and SNAP-tags
Abstract
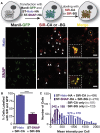
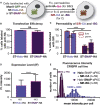
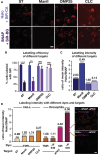
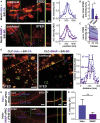
Super-resolution microscopy requires that subcellular structures are labeled with bright and photostable fluorophores, especially for live-cell imaging. Organic fluorophores may help here as they can yield more photons-by orders of magnitude-than fluorescent proteins. To achieve molecular specificity with organic fluorophores in live cells, self-labeling proteins are often used, with HaloTags and SNAP-tags being the most common. However, how these two different tagging systems compare with each other is unclear, especially for stimulated emission depletion (STED) microscopy, which is limited to a small repertoire of fluorophores in living cells. Herein, we compare the two labeling approaches in confocal and STED imaging using various proteins and two model systems. Strikingly, we find that the fluorescent signal can be up to 9-fold higher with HaloTags than with SNAP-tags when using far-red rhodamine derivatives. This result demonstrates that the labeling strategy matters and can greatly influence the duration of super-resolution imaging.
Keywords: HaloTag; SNAP-tag; STED; fluorophores; live-cell imaging; microscopy; nanoscopy; self-labeling proteins; super-resolution microscopy.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Conflict of interest statement
J.B. discloses financial interest in Bruker Corp. and Hamamatsu Photonics. The remaining authors declare no competing financial interests.
Figures


Similar articles
-
Green-Emitting Rhodamine Dyes for Vital Labeling of Cell Organelles Using STED Super-Resolution Microscopy.Chembiochem. 2019 Sep 2;20(17):2248-2254. doi: 10.1002/cbic.201900177. Epub 2019 May 21. Chembiochem. 2019. PMID: 31050112
-
Quantum-yield-optimized fluorophores for site-specific labeling and super-resolution imaging.J Am Chem Soc. 2011 Jun 1;133(21):8090-3. doi: 10.1021/ja200967z. Epub 2011 May 11. J Am Chem Soc. 2011. PMID: 21545135
-
Photoactivatable Large Stokes Shift Fluorophores for Multicolor Nanoscopy.J Am Chem Soc. 2023 Jan 25;145(3):1530-1534. doi: 10.1021/jacs.2c12567. Epub 2023 Jan 10. J Am Chem Soc. 2023. PMID: 36626161 Free PMC article.
-
Live-cell super-resolution imaging with synthetic fluorophores.Annu Rev Phys Chem. 2012;63:519-40. doi: 10.1146/annurev-physchem-032811-112012. Epub 2012 Jan 30. Annu Rev Phys Chem. 2012. PMID: 22404589 Review.
-
Strategies to maximize performance in STimulated Emission Depletion (STED) nanoscopy of biological specimens.Methods. 2020 Mar 1;174:27-41. doi: 10.1016/j.ymeth.2019.07.019. Epub 2019 Jul 22. Methods. 2020. PMID: 31344404 Free PMC article. Review.
Cited by
-
Advanced imaging and labelling methods to decipher brain cell organization and function.Nat Rev Neurosci. 2021 Apr;22(4):237-255. doi: 10.1038/s41583-021-00441-z. Epub 2021 Mar 12. Nat Rev Neurosci. 2021. PMID: 33712727 Review.
-
Beta HPV Deregulates Double-Strand Break Repair.Viruses. 2022 Apr 30;14(5):948. doi: 10.3390/v14050948. Viruses. 2022. PMID: 35632690 Free PMC article. Review.
-
Integration and Spatial Organization of Signaling by G Protein-Coupled Receptor Homo- and Heterodimers.Biomolecules. 2021 Dec 3;11(12):1828. doi: 10.3390/biom11121828. Biomolecules. 2021. PMID: 34944469 Free PMC article. Review.
-
Unraveling protein dynamics to understand the brain - the next molecular frontier.Mol Neurodegener. 2022 Jun 18;17(1):45. doi: 10.1186/s13024-022-00546-8. Mol Neurodegener. 2022. PMID: 35717317 Free PMC article. Review.
-
ULK1 forms distinct oligomeric states and nanoscopic structures during autophagy initiation.Sci Adv. 2023 Sep 29;9(39):eadh4094. doi: 10.1126/sciadv.adh4094. Epub 2023 Sep 29. Sci Adv. 2023. PMID: 37774021 Free PMC article.
References
-
- de Chaumont F., Dallongeville S., Chenouard N., Herve N., Pop S., Provoost T., Meas-Yedid V., Pankajakshan P., Lecomte T., Le Montagner Y. Icy: an open bioimage informatics platform for extended reproducible research. Nat. Meth. 2012;9:690–696. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

