Atomic force microscopy for single molecule characterisation of protein aggregation
- PMID: 30742801
- PMCID: PMC6420408
- DOI: 10.1016/j.abb.2019.02.001
Atomic force microscopy for single molecule characterisation of protein aggregation
Abstract
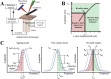
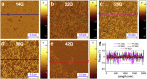
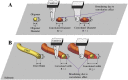
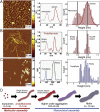
The development of atomic force microscopy (AFM) has opened up a wide range of novel opportunities in nanoscience and new modalities of observation in complex biological systems. AFM imaging has been widely employed to resolve the complex and heterogeneous conformational states involved in protein aggregation at the single molecule scale and shed light onto the molecular basis of a variety of human pathologies, including neurodegenerative disorders. The study of individual macromolecules at nanoscale, however, remains challenging, especially when fully quantitative information is required. In this review, we first discuss the principles of AFM with a special emphasis on the fundamental factors defining its sensitivity and accuracy. We then review the fundamental parameters and approaches to work at the limit of AFM resolution in order to perform single molecule statistical analysis of biomolecules and nanoscale protein aggregates. This single molecule statistical approach has proved to be powerful to unravel the molecular and hierarchical assembly of the misfolded species present transiently during protein aggregation, to visualise their dynamics at the nanoscale, as well to study the structural properties of amyloid-inspired functional nanomaterials.
Keywords: Amyloid; Atomic force microscopy; Biophysics; Protein aggregation; Resolution; Single molecule imaging.
Copyright © 2019. Published by Elsevier Inc.
Figures




Similar articles
-
AFM-Based Single Molecule Techniques: Unraveling the Amyloid Pathogenic Species.Curr Pharm Des. 2016;22(26):3950-70. doi: 10.2174/1381612822666160518141911. Curr Pharm Des. 2016. PMID: 27189600 Free PMC article. Review.
-
Atomic force microscopy as an imaging tool to study the bio/nonbio complexes.J Microsc. 2020 Dec;280(3):241-251. doi: 10.1111/jmi.12936. Epub 2020 Jun 17. J Microsc. 2020. PMID: 32519330
-
Atomic Force Microscopy for Protein Detection and Their Physicoсhemical Characterization.Int J Mol Sci. 2018 Apr 10;19(4):1142. doi: 10.3390/ijms19041142. Int J Mol Sci. 2018. PMID: 29642632 Free PMC article. Review.
-
Correlative Atomic Force and Single-Molecule Fluorescence Microscopy of Nucleoprotein Complexes.Methods Mol Biol. 2018;1814:339-359. doi: 10.1007/978-1-4939-8591-3_20. Methods Mol Biol. 2018. PMID: 29956242
-
Characterizing Individual Protein Aggregates by Infrared Nanospectroscopy and Atomic Force Microscopy.J Vis Exp. 2019 Sep 12;(151). doi: 10.3791/60108. J Vis Exp. 2019. PMID: 31566623
Cited by
-
Applications of Single-Molecule Vibrational Spectroscopic Techniques for the Structural Investigation of Amyloid Oligomers.Molecules. 2022 Sep 30;27(19):6448. doi: 10.3390/molecules27196448. Molecules. 2022. PMID: 36234985 Free PMC article. Review.
-
MAPT mutations associated with familial tauopathies lead to formation of conformationally distinct oligomers that have cross-seeding ability.Protein Sci. 2024 Sep;33(9):e5099. doi: 10.1002/pro.5099. Protein Sci. 2024. PMID: 39145409 Free PMC article.
-
Time-Resolved In Situ AFM Measurement of Growth Rates of Aβ40 Fibrils.Methods Mol Biol. 2023;2551:63-77. doi: 10.1007/978-1-0716-2597-2_6. Methods Mol Biol. 2023. PMID: 36310197
-
Single-Molecular Heteroamyloidosis of Human Islet Amyloid Polypeptide.Nano Lett. 2019 Sep 11;19(9):6535-6546. doi: 10.1021/acs.nanolett.9b02771. Epub 2019 Aug 29. Nano Lett. 2019. PMID: 31455083 Free PMC article.
-
MIRRAGGE - Minimum Information Required for Reproducible AGGregation Experiments.Front Mol Neurosci. 2020 Nov 27;13:582488. doi: 10.3389/fnmol.2020.582488. eCollection 2020. Front Mol Neurosci. 2020. PMID: 33328883 Free PMC article. Review.
References
-
- Binnig G., Quate C.F., Gerber C. Atomic force microscope. Phys. Rev. Lett. 1986;56:930–933. - PubMed
-
- Chang K.C., Chiang Y.W., Yang C.H., Liou J.W. Atomic force microscopy in biology and biomedicine. Tzu Chi Med. J. 2012;24:162–169.
-
- Variola F. Atomic force microscopy in biomaterials surface science. Phys. Chem. Chem. Phys. 2015;17:2950–2959. - PubMed
-
- Drolle E., Hane F., Lee B., Leonenko Z. Atomic force microscopy to study molecular mechanisms of amyloid fibril formation and toxicity in Alzheimer's disease. Drug Metab. Rev. 2014;46:207–223. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

