Influenza M2 protein regulates MAVS-mediated signaling pathway through interacting with MAVS and increasing ROS production
- PMID: 30741586
- PMCID: PMC6613841
- DOI: 10.1080/15548627.2019.1580089
Influenza M2 protein regulates MAVS-mediated signaling pathway through interacting with MAVS and increasing ROS production
Abstract
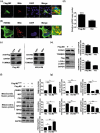
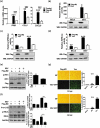
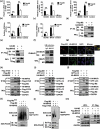
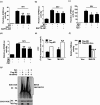
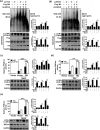
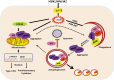
Influenza A virus can evade host innate immune response that is involved in several viral proteins with complicated mechanisms. To date, how influenza A M2 protein modulates the host innate immunity remains unclear. Herein, we showed that M2 protein colocalized and interacted with MAVS (mitochondrial antiviral signaling protein) on mitochondria, and positively regulated MAVS-mediated innate immunity. Further studies revealed that M2 induced reactive oxygen species (ROS) production that was required for activation of macroautophagy/autophagy and enhancement of MAVS signaling pathway. Importantly, the proton channel activity of M2 protein was demonstrated to be essential for ROS production and antagonizing the autophagy pathway to control MAVS aggregation, thereby enhancing MAVS signal activity. In conclusion, our studies provided novel insights into mechanisms of M2 protein in modulating host antiviral immunity and uncovered a new mechanism into biology and pathogenicity of influenza A virus. Abbreviations: AKT/PKB: AKT serine/threonine kinase; Apo: apocynin; ATG5: autophagy related 5; BAPTA-AM: 1,2-Bis(2-aminophenoxy) ethane-N,N,N',N'-tetraacetic acid tetrakis; BECN1: beclin 1; CARD: caspase recruitment domain; CCCP: carbonyl cyanide m-chlorophenylhydrazone; CQ: chloroquine; DCF: dichlorodihyd-rofluorescein; DPI: diphenyleneiodonium; DDX58: DExD/H-box helicase 58; eGFP: enhanced green fluorescent protein; EGTA: ethylene glycol-bis(2-aminoethylether)-N,N,N',N'-tetraacetic acid; ER: endoplasmic reticulum; hpi: hours post infection; IAV: influenza A virus; IFN: interferon; IP: immunoprecipitation; IRF3: interferon regulatory factor 3; ISRE: IFN-stimulated response elements; LIR: LC3-interacting region; MAP1LC3B/LC3B: microtubule associated protein 1 light chain 3 beta; MAVS: mitochondrial antiviral signaling protein; MMP: mitochondrial membrane potential; MOI, multiplicity of infection; mRFP: monomeric red fluorescent protein; MTOR: mechanistic target of rapamycin kinase; NC: negative control; NFKB/NF-κB: nuclear factor kappa B; PI3K: class I phosphoinositide 3-kinase; RLR: RIG-I-like-receptor; ROS: reactive oxygen species; SEV: sendai virus; TM: transmembrane; TMRM: tetramethylrhodamine methylester; VSV: vesicular stomatitis virus.
Keywords: Autophagy; MAVS aggregates; influenza M2 protein; innate immunity; ion channel activity.
Figures


Similar articles
-
Influenza A virus protein PB1-F2 impairs innate immunity by inducing mitophagy.Autophagy. 2021 Feb;17(2):496-511. doi: 10.1080/15548627.2020.1725375. Epub 2020 Feb 11. Autophagy. 2021. PMID: 32013669 Free PMC article.
-
BHRF1, a BCL2 viral homolog, disturbs mitochondrial dynamics and stimulates mitophagy to dampen type I IFN induction.Autophagy. 2021 Jun;17(6):1296-1315. doi: 10.1080/15548627.2020.1758416. Epub 2020 May 13. Autophagy. 2021. PMID: 32401605 Free PMC article.
-
Emerging views of mitophagy in immunity and autoimmune diseases.Autophagy. 2020 Jan;16(1):3-17. doi: 10.1080/15548627.2019.1603547. Epub 2019 Apr 21. Autophagy. 2020. PMID: 30951392 Free PMC article.
-
Mechanisms of MAVS regulation at the mitochondrial membrane.J Mol Biol. 2013 Dec 13;425(24):5009-19. doi: 10.1016/j.jmb.2013.10.007. Epub 2013 Oct 9. J Mol Biol. 2013. PMID: 24120683 Free PMC article. Review.
-
Activation of RIG-I-Mediated Antiviral Signaling Triggers Autophagy Through the MAVS-TRAF6-Beclin-1 Signaling Axis.Front Immunol. 2018 Sep 12;9:2096. doi: 10.3389/fimmu.2018.02096. eCollection 2018. Front Immunol. 2018. PMID: 30258449 Free PMC article. Review.
Cited by
-
Effect of natural products on host cell autophagy induced by Influenza A virus infection.Front Cell Infect Microbiol. 2024 Sep 30;14:1460604. doi: 10.3389/fcimb.2024.1460604. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39403204 Free PMC article. Review.
-
Mitochondrial Hyperactivity and Reactive Oxygen Species Drive Innate Immunity to the Yellow Fever Virus-17D Live-Attenuated Vaccine.bioRxiv [Preprint]. 2024 Sep 15:2024.09.04.611167. doi: 10.1101/2024.09.04.611167. bioRxiv. 2024. PMID: 39282299 Free PMC article. Preprint.
-
Reactive Oxygen Species Mechanisms that Regulate Protein-Protein Interactions in Cancer.Int J Mol Sci. 2024 Aug 27;25(17):9255. doi: 10.3390/ijms25179255. Int J Mol Sci. 2024. PMID: 39273204 Free PMC article. Review.
-
Pseudorabies virus infection triggers mitophagy to dampen the interferon response and promote viral replication.J Virol. 2024 Oct 22;98(10):e0104824. doi: 10.1128/jvi.01048-24. Epub 2024 Aug 30. J Virol. 2024. PMID: 39212384
-
The Hemagglutinin of Influenza A Virus Induces Ferroptosis to Facilitate Viral Replication.Adv Sci (Weinh). 2024 Oct;11(39):e2404365. doi: 10.1002/advs.202404365. Epub 2024 Aug 19. Adv Sci (Weinh). 2024. PMID: 39159143 Free PMC article.
References
-
- Chen C, Chi X, Bai Q, et al. Mechanisms underlying interferon-mediated host innate immunity during influenza A virus infection. Sheng Wu Gong Cheng Xue Bao. 2015;31(12):1671. - PubMed
-
- Mould JA, Li H-C, Dudlak CS, et al. Mechanism for proton conduction of the M2 ion channel of influenza A virus. J Biol Chem. 2000. March 24;275(12):8592–8599. PubMed PMID: BCI:BCI200000349462. - PubMed
-
- Pinto LH, Lamb RA.. Controlling influenza virus replication by inhibiting its proton channel. Mol Biosyst. 2007. January;3(1):18–23. PubMed PMID: WOS:000243429900013; English. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
