RNAs in the spliceosome: Insight from cryoEM structures
- PMID: 30729694
- PMCID: PMC6450755
- DOI: 10.1002/wrna.1523
RNAs in the spliceosome: Insight from cryoEM structures
Abstract
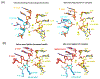
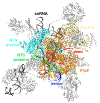
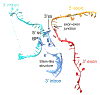
Pre-mRNA splicing is catalyzed by the spliceosome, a multimegadalton RNA-protein complex. The spliceosome undergoes dramatic compositional and conformational changes through the splicing cycle, forming at least 10 distinct complexes. Recent high-resolution cryoEM structures of various spliceosomal complexes revealed unprecedented details of this large molecular machine. This review highlights insight into the structure and function of the spliceosomal RNA components obtained from these new structures, with a focus on the yeast spliceosome. This article is categorized under: RNA Processing > Splicing Mechanisms RNA Structure and Dynamics > RNA Structure, Dynamics, and Chemistry RNA Interactions with Proteins and Other Molecules > RNA-Protein Complexes.
Keywords: RNA; cryoEM; spliceosome.
© 2019 Wiley Periodicals, Inc.
Figures

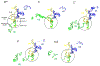

Similar articles
-
CryoEM structures of two spliceosomal complexes: starter and dessert at the spliceosome feast.Curr Opin Struct Biol. 2016 Feb;36:48-57. doi: 10.1016/j.sbi.2015.12.005. Epub 2016 Jan 21. Curr Opin Struct Biol. 2016. PMID: 26803803 Free PMC article. Review.
-
Cryo-EM structure of a human spliceosome activated for step 2 of splicing.Nature. 2017 Feb 16;542(7641):318-323. doi: 10.1038/nature21079. Epub 2017 Jan 11. Nature. 2017. PMID: 28076346
-
Mechanistic insights into precursor messenger RNA splicing by the spliceosome.Nat Rev Mol Cell Biol. 2017 Nov;18(11):655-670. doi: 10.1038/nrm.2017.86. Epub 2017 Sep 27. Nat Rev Mol Cell Biol. 2017. PMID: 28951565 Review.
-
Characterization of purified human Bact spliceosomal complexes reveals compositional and morphological changes during spliceosome activation and first step catalysis.RNA. 2010 Dec;16(12):2384-403. doi: 10.1261/rna.2456210. Epub 2010 Oct 27. RNA. 2010. PMID: 20980672 Free PMC article.
-
DEAH-Box RNA Helicases in Pre-mRNA Splicing.Trends Biochem Sci. 2021 Mar;46(3):225-238. doi: 10.1016/j.tibs.2020.10.006. Epub 2020 Nov 30. Trends Biochem Sci. 2021. PMID: 33272784 Free PMC article. Review.
Cited by
-
SPF45/RBM17-dependent, but not U2AF-dependent, splicing in a distinct subset of human short introns.Nat Commun. 2021 Aug 13;12(1):4910. doi: 10.1038/s41467-021-24879-y. Nat Commun. 2021. PMID: 34389706 Free PMC article.
-
Unique and Repeated Stwintrons (Spliceosomal Twin Introns) in the Hypoxylaceae.J Fungi (Basel). 2022 Apr 13;8(4):397. doi: 10.3390/jof8040397. J Fungi (Basel). 2022. PMID: 35448628 Free PMC article.
-
A unified mechanism for intron and exon definition and back-splicing.Nature. 2019 Sep;573(7774):375-380. doi: 10.1038/s41586-019-1523-6. Epub 2019 Sep 4. Nature. 2019. PMID: 31485080 Free PMC article.
-
Transcript-specific determinants of pre-mRNA splicing revealed through in vivo kinetic analyses of the 1st and 2nd chemical steps.Mol Cell. 2022 Aug 18;82(16):2967-2981.e6. doi: 10.1016/j.molcel.2022.06.020. Epub 2022 Jul 12. Mol Cell. 2022. PMID: 35830855 Free PMC article.
-
Monovalent metal ion binding promotes the first transesterification reaction in the spliceosome.Nat Commun. 2023 Dec 20;14(1):8482. doi: 10.1038/s41467-023-44174-2. Nat Commun. 2023. PMID: 38123540 Free PMC article.
References
-
- Mattick JS & Gagen MJ The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms. Mol Biol Evol 18, 1611–30 (2001). - PubMed
-
- Venables JP Alternative splicing in the testes. Curr Opin Genet Dev 12, 615–19 (2002). - PubMed
-
- Lopez AJ Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu Rev Genet 32, 279–305 (1998). - PubMed
-
- Grabowski PJ & Black DL Alternative RNA splicing in the nervous system. Prog Neurobiol 65, 289–308 (2001). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

