MAPK modulation of yeast pheromone signaling output and the role of phosphorylation sites in the scaffold protein Ste5
- PMID: 30726174
- PMCID: PMC6589907
- DOI: 10.1091/mbc.E18-12-0793
MAPK modulation of yeast pheromone signaling output and the role of phosphorylation sites in the scaffold protein Ste5
Abstract
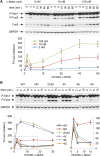
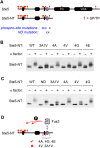
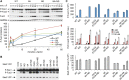
Mitogen-activated protein kinases (MAPKs) mediate numerous eukaryotic signaling responses. They also can modulate their own signaling output via positive or negative feedback loops. In the yeast pheromone response pathway, the MAPK Fus3 triggers negative feedback that dampens its own activity. One target of this feedback is Ste5, a scaffold protein that promotes Fus3 activation. Binding of Fus3 to a docking motif (D motif) in Ste5 causes signal dampening, which was proposed to involve a central cluster of phosphorylation sites in Ste5. Here, we reanalyzed the role of these central sites. Contrary to prior claims, phosphorylation-mimicking mutations at these sites did not impair signaling. Also, the hyperactive signaling previously observed when these sites were mutated to nonphosphorylatable residues arose from their replacement with valine residues and was not observed with other substitutes. Instead, a cluster of N-terminal sites in Ste5, not the central sites, is required for the rapid dampening of initial responses. Further results suggest that the role of the Fus3 D motif is most simply explained by a tethering effect that promotes Ste5 phosphorylation, rather than an allosteric effect proposed to regulate Fus3 activity. These findings substantially revise our understanding of how MAPK feedback attenuates scaffold-mediated signaling in this model pathway.
Figures

Similar articles
-
Control of MAPK signaling specificity by a conserved residue in the MEK-binding domain of the yeast scaffold protein Ste5.Curr Genet. 2006 Jun;49(6):351-63. doi: 10.1007/s00294-006-0061-6. Epub 2006 Feb 4. Curr Genet. 2006. PMID: 16463042
-
Persistent activation by constitutive Ste7 promotes Kss1-mediated invasive growth but fails to support Fus3-dependent mating in yeast.Mol Cell Biol. 2004 Oct;24(20):9221-38. doi: 10.1128/MCB.24.20.9221-9238.2004. Mol Cell Biol. 2004. PMID: 15456892 Free PMC article.
-
Mitogen-activated protein kinases with distinct requirements for Ste5 scaffolding influence signaling specificity in Saccharomyces cerevisiae.Mol Cell Biol. 2005 Mar;25(5):1793-803. doi: 10.1128/MCB.25.5.1793-1803.2005. Mol Cell Biol. 2005. PMID: 15713635 Free PMC article.
-
Pheromone signaling pathways in yeast.Sci STKE. 2006 Dec 5;2006(364):cm6. doi: 10.1126/stke.3642006cm6. Sci STKE. 2006. PMID: 17148787 Review.
-
Pheromone response, mating and cell biology.Curr Opin Microbiol. 2000 Dec;3(6):573-81. doi: 10.1016/s1369-5274(00)00143-0. Curr Opin Microbiol. 2000. PMID: 11121776 Review.
Cited by
-
The histone H2B Arg95 residue efficiently recruits the transcription factor Spt16 to mediate Ste5 expression of the pheromone response pathway.Sci Rep. 2023 Jun 22;13(1):10189. doi: 10.1038/s41598-023-37339-y. Sci Rep. 2023. PMID: 37349401 Free PMC article.
-
The Mitogen-Activated Protein Kinase Slt2 Promotes Asymmetric Cell Cycle Arrest and Reduces TORC1-Sch9 Signaling in Yeast Lacking the Protein Phosphatase Ptc1.Microbiol Spectr. 2023 Jun 15;11(3):e0524922. doi: 10.1128/spectrum.05249-22. Epub 2023 Apr 12. Microbiol Spectr. 2023. PMID: 37042757 Free PMC article.
-
The Paxillin MoPax1 Activates Mitogen-Activated Protein (MAP) Kinase Signaling Pathways and Autophagy through MAP Kinase Activator MoMka1 during Appressorium-Mediated Plant Infection by the Rice Blast Fungus Magnaporthe oryzae.mBio. 2022 Dec 20;13(6):e0221822. doi: 10.1128/mbio.02218-22. Epub 2022 Oct 31. mBio. 2022. PMID: 36314807 Free PMC article.
-
The histone H2B Arg95 residue links the pheromone response pathway to rapamycin-induced G1 arrest in yeast.Sci Rep. 2022 Jun 15;12(1):10023. doi: 10.1038/s41598-022-14053-9. Sci Rep. 2022. PMID: 35705668 Free PMC article.
-
Mechanical stress impairs pheromone signaling via Pkc1-mediated regulation of the MAPK scaffold Ste5.J Cell Biol. 2019 Sep 2;218(9):3117-3133. doi: 10.1083/jcb.201808161. Epub 2019 Jul 17. J Cell Biol. 2019. PMID: 31315942 Free PMC article.
References
-
- Bhattacharyya RP, Remenyi A, Good MC, Bashor CJ, Falick AM, Lim WA. (2006). The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science , 822–826. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

