Oleic acid induces migration through a FFAR1/4, EGFR and AKT-dependent pathway in breast cancer cells
- PMID: 30721135
- PMCID: PMC6410766
- DOI: 10.1530/EC-18-0543
Oleic acid induces migration through a FFAR1/4, EGFR and AKT-dependent pathway in breast cancer cells
Abstract
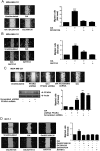
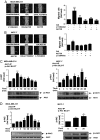
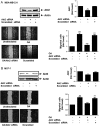
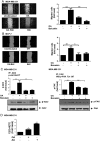
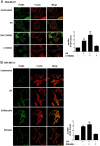
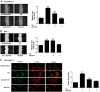
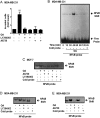
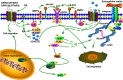
Free fatty acids (FFAs) are an energy source, and induce activation of signal transduction pathways that mediate several biological processes. In breast cancer cells, oleic acid (OA) induces proliferation, matrix metalloproteinase-9 (MMP-9) secretion, migration and invasion. However, the signal transduction pathways that mediate migration and invasion induced by OA in breast cancer cells have not been studied in detail. We demonstrate here that FFAR1 and FFAR4 mediate migration induced by OA in MDA-MB-231 and MCF-7 breast cancer cells. Moreover, OA induces migration, invasion, AKT1 and AKT2 activation, 12-LOX secretion and an increase of NFκB-DNA binding activity in breast cancer cells. Cell migration requires FFAR1, FFAR4, EGFR, AKT and PI3K activity, whereas invasion is mediated though a PI3K/Akt-dependent pathway. Furthermore, OA promotes relocalization of paxillin to focal contacts and it requires PI3K and EGFR activity, whereas NFκB-DNA binding activity requires PI3K and AKT activity.
Keywords: 12-LOX; breast cancer; invasion; migration; oleic acid (OA).
Figures
Similar articles
-
Linoleic acid induces migration and invasion through FFAR4- and PI3K-/Akt-dependent pathway in MDA-MB-231 breast cancer cells.Med Oncol. 2017 Jun;34(6):111. doi: 10.1007/s12032-017-0969-3. Epub 2017 Apr 29. Med Oncol. 2017. PMID: 28456993
-
Arachidonic acid promotes migration and invasion through a PI3K/Akt-dependent pathway in MDA-MB-231 breast cancer cells.Prostaglandins Leukot Essent Fatty Acids. 2014 May;90(5):169-77. doi: 10.1016/j.plefa.2014.01.007. Epub 2014 Feb 12. Prostaglandins Leukot Essent Fatty Acids. 2014. PMID: 24565443
-
Oleic acid promotes MMP-9 secretion and invasion in breast cancer cells.Clin Exp Metastasis. 2010 Oct;27(7):505-15. doi: 10.1007/s10585-010-9340-1. Epub 2010 Jul 9. Clin Exp Metastasis. 2010. PMID: 20617371
-
Osteopontin: it's role in regulation of cell motility and nuclear factor kappa B-mediated urokinase type plasminogen activator expression.IUBMB Life. 2005 Jun;57(6):441-7. doi: 10.1080/15216540500159424. IUBMB Life. 2005. PMID: 16012053 Review.
-
For Better or Worse: FFAR1 and FFAR4 Signaling in Cancer and Diabetes.Mol Pharmacol. 2016 Dec;90(6):738-743. doi: 10.1124/mol.116.105932. Epub 2016 Aug 31. Mol Pharmacol. 2016. PMID: 27582526 Review.
Cited by
-
Oleic Acid Exhibits Anti-Proliferative and Anti-Invasive Activities via the PTEN/AKT/mTOR Pathway in Endometrial Cancer.Cancers (Basel). 2023 Nov 14;15(22):5407. doi: 10.3390/cancers15225407. Cancers (Basel). 2023. PMID: 38001668 Free PMC article.
-
Distinct functions of AKT isoforms in breast cancer: a comprehensive review.Cell Commun Signal. 2019 Nov 21;17(1):154. doi: 10.1186/s12964-019-0450-3. Cell Commun Signal. 2019. PMID: 31752925 Free PMC article. Review.
-
Role of adipocyte browning in prostate and breast tumor microenvironment.Tzu Chi Med J. 2022 Jun 27;34(4):359-366. doi: 10.4103/tcmj.tcmj_62_22. eCollection 2022 Oct-Dec. Tzu Chi Med J. 2022. PMID: 36578640 Free PMC article. Review.
-
Nobiletin Inhibits Cell Viability via the SRC/AKT/STAT3/YY1AP1 Pathway in Human Renal Carcinoma Cells.Front Pharmacol. 2019 Jul 9;10:690. doi: 10.3389/fphar.2019.00690. eCollection 2019. Front Pharmacol. 2019. PMID: 31354472 Free PMC article.
-
Oleate Promotes Triple-Negative Breast Cancer Cell Migration by Enhancing Filopodia Formation through a PLD/Cdc42-Dependent Pathway.Int J Mol Sci. 2024 Apr 2;25(7):3956. doi: 10.3390/ijms25073956. Int J Mol Sci. 2024. PMID: 38612766 Free PMC article.
References
-
- Rossini A, Zanobbio L, Sfondrini L, Cavalleri A, Secreto G, Morelli D, Palazzo M, Sommariva M, Tagliabue E, Rumio C, et al Influence of fatty acid-free diet on mammary tumor development and growth rate in HER-2/Neu transgenic mice. Journal of Cellular Physiology 2013. 228 242–249. (10.1002/jcp.24130) - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

