Maternal regulation of chromosomal imprinting in animals
- PMID: 30719566
- PMCID: PMC6536480
- DOI: 10.1007/s00412-018-00690-5
Maternal regulation of chromosomal imprinting in animals
Abstract
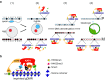
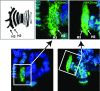
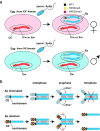
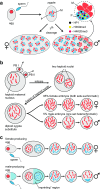
Chromosomal imprinting requires an epigenetic system that "imprints" one of the two parental chromosomes such that it results in a heritable (cell-to-cell) change in behavior of the "imprinted" chromosome. Imprinting takes place when the parental genomes are separate, which occurs during gamete formation in the respective germ-lines and post-fertilization during the period when the parental pro-nuclei lie separately within the ooplasm of the zygote. In the mouse, chromosomal imprinting is regulated by germ-line specific DNA methylation. But the methylation machinery in the respective germ-lines does not discriminate between imprinted and non-imprinted regions. As a consequence, the mouse oocyte nucleus contains over a thousand oocyte-specific germ-line differentially methylated regions (gDMRs). Upon fertilization, the sperm provides a few hundred sperm-specific gDMRs of its own. Combined, there are around 1600 imprinted and non-imprinted gDMRs in the pro-nuclei of the newly fertilized zygote. It is a remarkable fact that beginning in the maternal ooplasm, there are mechanisms that manage to preserve DNA methylation at ~ 26 known imprinted gDMRs in the face of the ongoing genome-wide DNA de-methylation that characterizes pre-implantation development. Specificity is achieved through the binding of KRAB-zinc finger proteins to their cognate recognition sequences within the gDMRs of imprinted genes. This in turn nucleates the assembly of localized heterochromatin-like complexes that preserve methylation at imprinted gDMRs through recruitment of the maintenance methyl transferase Dnmt1. These studies have shown that a germ-line imprint may cause parent-of-origin-specific behavior only if "licensed" by mechanisms that operate post-fertilization. Study of the germ-line and post-fertilization contributions to the imprinting of chromosomes in classical insect systems (Coccidae and Sciaridae) show that the ooplasm is the likely site where imprinting takes place. By comparing molecular and genetic studies across these three species, we suggest that mechanisms which operate post-fertilization play a key role in chromosomal imprinting phenomena in animals and conserved components of heterochromatin are shared by these mechanisms.
Keywords: Chromosomal imprinting; Epigenetics; Genomic imprinting; Germ-line differentially methylated regions; H3K9me3:HP1:H4K20me3 pathway; Heterochromatin; Mus musculus; Non-coding RNA; Parent-of-origin effects; Plannococcus citri; Sciara coprophila.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Similar articles
-
Imprinting of the mouse Igf2r gene depends on an intronic CpG island.Mol Cell Endocrinol. 1998 May 25;140(1-2):9-14. doi: 10.1016/s0303-7207(98)00022-7. Mol Cell Endocrinol. 1998. PMID: 9722161 Review.
-
The loss of imprinted DNA methylation in mouse blastocysts is inflicted to a similar extent by in vitro follicle culture and ovulation induction.Mol Hum Reprod. 2016 Jun;22(6):427-41. doi: 10.1093/molehr/gaw013. Epub 2016 Feb 7. Mol Hum Reprod. 2016. PMID: 26908643
-
Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks.PLoS Genet. 2012 Jan;8(1):e1002440. doi: 10.1371/journal.pgen.1002440. Epub 2012 Jan 5. PLoS Genet. 2012. PMID: 22242016 Free PMC article.
-
The specification of imprints in mammals.Heredity (Edinb). 2014 Aug;113(2):176-83. doi: 10.1038/hdy.2014.54. Epub 2014 Jun 18. Heredity (Edinb). 2014. PMID: 24939713 Free PMC article. Review.
-
Genomic imprinting is a parental effect established in mammalian germ cells.Curr Top Dev Biol. 2013;102:35-59. doi: 10.1016/B978-0-12-416024-8.00002-7. Curr Top Dev Biol. 2013. PMID: 23287029 Review.
Cited by
-
H3K9 and H4K20 methyltransferases are directly involved in the heterochromatinization of the paternal chromosomes in male Planococcus citri embryos.Chromosoma. 2023 Nov;132(4):317-328. doi: 10.1007/s00412-023-00809-3. Epub 2023 Sep 12. Chromosoma. 2023. PMID: 37700063
-
On the relations of phase separation and Hi-C maps to epigenetics.R Soc Open Sci. 2020 Feb 26;7(2):191976. doi: 10.1098/rsos.191976. eCollection 2020 Feb. R Soc Open Sci. 2020. PMID: 32257349 Free PMC article.
-
Trans Species RNA Activity: Sperm RNA of the Father of an Autistic Child Programs Glial Cells and Behavioral Disorders in Mice.Biomolecules. 2024 Feb 7;14(2):201. doi: 10.3390/biom14020201. Biomolecules. 2024. PMID: 38397438 Free PMC article.
-
Biology and Physics of Heterochromatin-Like Domains/Complexes.Cells. 2020 Aug 11;9(8):1881. doi: 10.3390/cells9081881. Cells. 2020. PMID: 32796726 Free PMC article. Review.
-
Increased copy number of imprinted genes in the chromosomal region 20q11-q13.32 is associated with resistance to antitumor agents in cancer cell lines.Clin Epigenetics. 2022 Dec 2;14(1):161. doi: 10.1186/s13148-022-01368-7. Clin Epigenetics. 2022. PMID: 36461044 Free PMC article.
References
-
- Anvar Z, Cammisa M, Riso V, Baglivo I, Kukreja H, Sparago A, Girardot M, Lad S, de Feis I, Cerrato F, Angelini C, Feil R, Pedone PV, Grimaldi G, Riccio A. ZFP57 recognizes multiple and closely spaced sequence motif variants to maintain repressive epigenetic marks in mouse embryonic stem cells. Nucleic Acids Res. 2016;44:1118–1132. doi: 10.1093/nar/gkv1059. - DOI - PMC - PubMed
-
- Arney KL, Bao S, Bannister AJ, Kouzarides T, Surani MA. Histone methylation defines epigenetic asymmetry in the mouse zygote. Int J Dev Biol. 2002;46:317–320. - PubMed
-
- Berry RO. Chromosome behavior in the germ cells and development of the gonads in Sciara ocellaris. J Morphol. 1941;68:547–583. doi: 10.1002/jmor.1050680307. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

