CD4 receptor diversity in chimpanzees protects against SIV infection
- PMID: 30718403
- PMCID: PMC6386711
- DOI: 10.1073/pnas.1821197116
CD4 receptor diversity in chimpanzees protects against SIV infection
Abstract
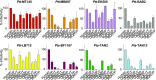
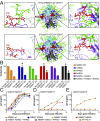
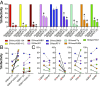
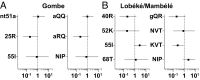
Human and simian immunodeficiency viruses (HIV/SIVs) use CD4 as the primary receptor to enter target cells. Here, we show that the chimpanzee CD4 is highly polymorphic, with nine coding variants present in wild populations, and that this diversity interferes with SIV envelope (Env)-CD4 interactions. Testing the replication fitness of SIVcpz strains in CD4+ T cells from captive chimpanzees, we found that certain viruses were unable to infect cells from certain hosts. These differences were recapitulated in CD4 transfection assays, which revealed a strong association between CD4 genotypes and SIVcpz infection phenotypes. The most striking differences were observed for three substitutions (Q25R, Q40R, and P68T), with P68T generating a second N-linked glycosylation site (N66) in addition to an invariant N32 encoded by all chimpanzee CD4 alleles. In silico modeling and site-directed mutagenesis identified charged residues at the CD4-Env interface and clashes between CD4- and Env-encoded glycans as mechanisms of inhibition. CD4 polymorphisms also reduced Env-mediated cell entry of monkey SIVs, which was dependent on at least one D1 domain glycan. CD4 allele frequencies varied among wild chimpanzees, with high diversity in all but the western subspecies, which appeared to have undergone a selective sweep. One allele was associated with lower SIVcpz prevalence rates in the wild. These results indicate that substitutions in the D1 domain of the chimpanzee CD4 can prevent SIV cell entry. Although some SIVcpz strains have adapted to utilize these variants, CD4 diversity is maintained, protecting chimpanzees against infection with SIVcpz and other SIVs to which they are exposed.
Keywords: CD4; SIV; chimpanzee; envelope glycoprotein; glycan restriction.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
A glycan shield on chimpanzee CD4 protects against infection by primate lentiviruses (HIV/SIV).Proc Natl Acad Sci U S A. 2019 Jun 4;116(23):11460-11469. doi: 10.1073/pnas.1813909116. Epub 2019 May 21. Proc Natl Acad Sci U S A. 2019. PMID: 31113887 Free PMC article.
-
CD4 receptor diversity represents an ancient protection mechanism against primate lentiviruses.Proc Natl Acad Sci U S A. 2021 Mar 30;118(13):e2025914118. doi: 10.1073/pnas.2025914118. Proc Natl Acad Sci U S A. 2021. PMID: 33771926 Free PMC article.
-
Neutralization properties of simian immunodeficiency viruses infecting chimpanzees and gorillas.mBio. 2015 Apr 21;6(2):e00296-15. doi: 10.1128/mBio.00296-15. mBio. 2015. PMID: 25900654 Free PMC article.
-
[SIV as a source of HIV. On the origin of human immunodeficiency viruses from non-human primates].Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2004 Jul;47(7):680-4. doi: 10.1007/s00103-004-0862-z. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2004. PMID: 15254823 Review. German.
-
The history of SIVS and AIDS: epidemiology, phylogeny and biology of isolates from naturally SIV infected non-human primates (NHP) in Africa.Front Biosci. 2004 Jan 1;9:225-54. doi: 10.2741/1154. Front Biosci. 2004. PMID: 14766362 Review.
Cited by
-
A survey of the adaptive immune genes of the polka-dot batfish Ogcocephalus cubifrons.BMC Immunol. 2023 Jul 21;24(1):20. doi: 10.1186/s12865-023-00557-0. BMC Immunol. 2023. PMID: 37480016 Free PMC article.
-
Widespread extinctions of co-diversified primate gut bacterial symbionts from humans.Nat Microbiol. 2023 Jun;8(6):1039-1050. doi: 10.1038/s41564-023-01388-w. Epub 2023 May 11. Nat Microbiol. 2023. PMID: 37169918 Free PMC article.
-
Comparative physiological anthropogeny: exploring molecular underpinnings of distinctly human phenotypes.Physiol Rev. 2023 Jul 1;103(3):2171-2229. doi: 10.1152/physrev.00040.2021. Epub 2023 Jan 5. Physiol Rev. 2023. PMID: 36603157 Free PMC article. Review.
-
Genetic adaptations to SIV across chimpanzee populations.PLoS Genet. 2022 Aug 25;18(8):e1010337. doi: 10.1371/journal.pgen.1010337. eCollection 2022 Aug. PLoS Genet. 2022. PMID: 36007015 Free PMC article.
-
The Gombe Ecosystem Health Project: 16 years of program evolution and lessons learned.Am J Primatol. 2022 May;84(4-5):e23300. doi: 10.1002/ajp.23300. Epub 2021 Jul 5. Am J Primatol. 2022. PMID: 34223656 Free PMC article. Review.
References
-
- Peeters M, Ma D, Liegeois F, Apetrei C. Simian immunodeficiency virus infections in the wild. In: Ansari A, Silvestri G, editors. Natural Hosts of SIV. Elsevier; New York: 2014. pp. 37–67.
-
- Van Heuverswyn F, et al. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature. 2006;444:164. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- UM1 AI100645/AI/NIAID NIH HHS/United States
- T32 AI007632/AI/NIAID NIH HHS/United States
- R37 AI050529/AI/NIAID NIH HHS/United States
- 352417/CAPMC/ CIHR/Canada
- R37 AI150590/AI/NIAID NIH HHS/United States
- R01 AI120810/AI/NIAID NIH HHS/United States
- R01 AI091595/AI/NIAID NIH HHS/United States
- P01 AI131251/AI/NIAID NIH HHS/United States
- R01 AI050529/AI/NIAID NIH HHS/United States
- R01 AI058715/AI/NIAID NIH HHS/United States
- P51 OD011133/OD/NIH HHS/United States
- R61 AI133696/AI/NIAID NIH HHS/United States
- UM1 AI126620/AI/NIAID NIH HHS/United States
- P30 AI045008/AI/NIAID NIH HHS/United States
- P51 RR000165/RR/NCRR NIH HHS/United States
- R01 AI131331/AI/NIAID NIH HHS/United States
- R56 AI091516/AI/NIAID NIH HHS/United States
- R33 AI133696/AI/NIAID NIH HHS/United States
- UM1 AI144371/AI/NIAID NIH HHS/United States
- P51 RR013986/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

