Mesoscale modeling reveals formation of an epigenetically driven HOXC gene hub
- PMID: 30718394
- PMCID: PMC6421463
- DOI: 10.1073/pnas.1816424116
Mesoscale modeling reveals formation of an epigenetically driven HOXC gene hub
Abstract
Gene expression is orchestrated at the structural level by nucleosome positioning, histone tail acetylation, and linker histone (LH) binding. Here, we integrate available data on nucleosome positioning, nucleosome-free regions (NFRs), acetylation islands, and LH binding sites to "fold" in silico the 55-kb HOXC gene cluster and investigate the role of each feature on the gene's folding. The gene cluster spontaneously forms a dynamic connection hub, characterized by hierarchical loops which accommodate multiple contacts simultaneously and decrease the average distance between promoters by ∼100 nm. Contact probability matrices exhibit "stripes" near promoter regions, a feature associated with transcriptional regulation. Interestingly, while LH proteins alone decrease long-range contacts and acetylation alone increases transient contacts, combined LH and acetylation produce long-range contacts. Thus, our work emphasizes how chromatin architecture is coordinated strongly by epigenetic factors and opens the way for nucleosome resolution models incorporating epigenetic modifications to understand and predict gene activity.
Keywords: chromatin folding; chromatin loop domains; chromatin modeling; contact hub; gene structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
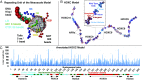


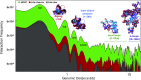

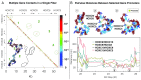
Comment in
-
Inner workings of gene folding.Proc Natl Acad Sci U S A. 2019 Mar 12;116(11):4774-4775. doi: 10.1073/pnas.1900875116. Epub 2019 Feb 22. Proc Natl Acad Sci U S A. 2019. PMID: 30796189 Free PMC article. No abstract available.
Similar articles
-
Nucleosome Clutches are Regulated by Chromatin Internal Parameters.J Mol Biol. 2021 Mar 19;433(6):166701. doi: 10.1016/j.jmb.2020.11.001. Epub 2020 Nov 9. J Mol Biol. 2021. PMID: 33181171 Free PMC article.
-
Chromatin Fiber Folding Directed by Cooperative Histone Tail Acetylation and Linker Histone Binding.Biophys J. 2018 May 22;114(10):2376-2385. doi: 10.1016/j.bpj.2018.03.008. Epub 2018 Apr 11. Biophys J. 2018. PMID: 29655483 Free PMC article.
-
Chromatin looping and the probability of transcription.Trends Genet. 2006 Apr;22(4):197-202. doi: 10.1016/j.tig.2006.02.004. Epub 2006 Feb 21. Trends Genet. 2006. PMID: 16494964 Review.
-
Reconstitution of nucleosome positioning, remodeling, histone acetylation, and transcriptional activation on the PHO5 promoter.J Biol Chem. 2002 Aug 23;277(34):31038-47. doi: 10.1074/jbc.M204662200. Epub 2002 Jun 11. J Biol Chem. 2002. PMID: 12060664
-
Epigenetic regulation in chondrogenesis.Acta Med Okayama. 2010 Jun;64(3):155-61. doi: 10.18926/AMO/40007. Acta Med Okayama. 2010. PMID: 20596126 Review.
Cited by
-
Genome modeling: From chromatin fibers to genes.Curr Opin Struct Biol. 2023 Feb;78:102506. doi: 10.1016/j.sbi.2022.102506. Epub 2022 Dec 26. Curr Opin Struct Biol. 2023. PMID: 36577295 Free PMC article. Review.
-
Heterogeneous interactions and polymer entropy decide organization and dynamics of chromatin domains.Biophys J. 2022 Jul 19;121(14):2794-2812. doi: 10.1016/j.bpj.2022.06.008. Epub 2022 Jun 6. Biophys J. 2022. PMID: 35672951 Free PMC article.
-
Integrative Modeling of 3D Genome Organization by Bayesian Molecular Dynamics Simulations with Hi-C Metainference.Methods Mol Biol. 2025;2856:309-324. doi: 10.1007/978-1-0716-4136-1_19. Methods Mol Biol. 2025. PMID: 39283461
-
Mesoscale Liquid Model of Chromatin Recapitulates Nuclear Order of Eukaryotes.Biophys J. 2020 May 5;118(9):2130-2140. doi: 10.1016/j.bpj.2019.09.013. Epub 2019 Sep 17. Biophys J. 2020. PMID: 31623887 Free PMC article.
-
Predicting scale-dependent chromatin polymer properties from systematic coarse-graining.Nat Commun. 2023 Jul 11;14(1):4108. doi: 10.1038/s41467-023-39907-2. Nat Commun. 2023. PMID: 37433821 Free PMC article.
References
-
- Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Gen. 2007;39:311–318. - PubMed
-
- Finch JT, et al. Structure of nucleosome core particles of chromatin. Nature. 1977;269:29–36. - PubMed
-
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

