Meta-omics data and collection objects (MOD-CO): a conceptual schema and data model for processing sample data in meta-omics research
- PMID: 30715273
- PMCID: PMC6354027
- DOI: 10.1093/database/baz002
Meta-omics data and collection objects (MOD-CO): a conceptual schema and data model for processing sample data in meta-omics research
Abstract
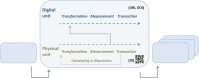
With the advent of advanced molecular meta-omics techniques and methods, a new era commenced for analysing and characterizing historic collection specimens, as well as recently collected environmental samples. Nucleic acid and protein sequencing-based analyses are increasingly applied to determine the origin, identity and traits of environmental (biological) objects and organisms. In this context, the need for new data structures is evident and former approaches for data processing need to be expanded according to the new meta-omics techniques and operational standards. Existing schemas and community standards in the biodiversity and molecular domain concentrate on terms important for data exchange and publication. Detailed operational aspects of origin and laboratory as well as object and data management issues are frequently neglected. Meta-omics Data and Collection Objects (MOD-CO) has therefore been set up as a new schema for meta-omics research, with a hierarchical organization of the concepts describing collection samples, as well as products and data objects being generated during operational workflows. It is focussed on object trait descriptions as well as on operational aspects and thereby may serve as a backbone for R&D laboratory information management systems with functions of an electronic laboratory notebook. The schema in its current version 1.0 includes 653 concepts and 1810 predefined concept values, being equivalent to descriptors and descriptor states, respectively. It is published in several representations, like a Semantic Media Wiki publication with 2463 interlinked Wiki pages for concepts and concept values, being grouped in 37 concept collections and subcollections. The SQL database application DiversityDescriptions, a generic tool for maintaining descriptive data and schemas, has been applied for setting up and testing MOD-CO and for concept mapping on elements of corresponding schemas.
Figures


Similar articles
-
FAIR digital objects in environmental and life sciences should comprise workflow operation design data and method information for repeatability of study setups and reproducibility of results.Database (Oxford). 2020 Jan 1;2020:baaa059. doi: 10.1093/database/baaa059. Database (Oxford). 2020. PMID: 32815545 Free PMC article.
-
Omics data management and annotation.Methods Mol Biol. 2011;719:71-96. doi: 10.1007/978-1-61779-027-0_3. Methods Mol Biol. 2011. PMID: 21370079
-
Toward a view-oriented approach for aligning RDF-based biomedical repositories.Methods Inf Med. 2015;54(1):50-5. doi: 10.3414/ME13-02-0020. Epub 2014 Apr 29. Methods Inf Med. 2015. PMID: 24777240
-
Translational Metabolomics of Head Injury: Exploring Dysfunctional Cerebral Metabolism with Ex Vivo NMR Spectroscopy-Based Metabolite Quantification.In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. PMID: 26269925 Free Books & Documents. Review.
-
Extension of research data repository system to support direct compute access to biomedical datasets: enhancing Dataverse to support large datasets.Ann N Y Acad Sci. 2017 Jan;1387(1):95-104. doi: 10.1111/nyas.13272. Epub 2016 Nov 10. Ann N Y Acad Sci. 2017. PMID: 27862010 Free PMC article. Review.
Cited by
-
FAIR digital objects in environmental and life sciences should comprise workflow operation design data and method information for repeatability of study setups and reproducibility of results.Database (Oxford). 2020 Jan 1;2020:baaa059. doi: 10.1093/database/baaa059. Database (Oxford). 2020. PMID: 32815545 Free PMC article.
-
GenoSurf: metadata driven semantic search system for integrated genomic datasets.Database (Oxford). 2019 Jan 1;2019:baz132. doi: 10.1093/database/baz132. Database (Oxford). 2019. PMID: 31820804 Free PMC article.
-
The archives are half-empty: an assessment of the availability of microbial community sequencing data.Commun Biol. 2020 Aug 28;3(1):474. doi: 10.1038/s42003-020-01204-9. Commun Biol. 2020. PMID: 32859925 Free PMC article.
-
Current Approaches for Advancement in Understanding the Molecular Mechanisms of Mycotoxin Biosynthesis.Int J Mol Sci. 2021 Jul 23;22(15):7878. doi: 10.3390/ijms22157878. Int J Mol Sci. 2021. PMID: 34360643 Free PMC article. Review.
References
-
- Wallenstein M.D., Haddix M.L., Ayres E. et al. (2013) Litter chemistry changes more rapidly when decomposed at home but converges during decomposition-transformation. Soil Biol. Biochem., 57, 311–319.
-
- Cusack D.F., Silver W.L., Torn M.S. et al. (2011) Changes in microbial community characteristics and soil organic matter with nitrogen additions in two tropical forests. Ecology, 92, 621–632. - PubMed
-
- Peršoh D. (2015) Plant-associated fungal communities in the light of meta-omics. Fungal Divers., 75, 1–25.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

