Adenosine deaminase acting on RNA (ADAR1), a suppressor of double-stranded RNA-triggered innate immune responses
- PMID: 30710018
- PMCID: PMC6364763
- DOI: 10.1074/jbc.TM118.004166
Adenosine deaminase acting on RNA (ADAR1), a suppressor of double-stranded RNA-triggered innate immune responses
Abstract
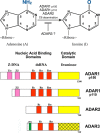
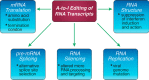
Herbert "Herb" Tabor, who celebrated his 100th birthday this past year, served the Journal of Biological Chemistry as a member of the Editorial Board beginning in 1961, as an Associate Editor, and as Editor-in-Chief for 40 years, from 1971 until 2010. Among the many discoveries in biological chemistry during this period was the identification of RNA modification by C6 deamination of adenosine (A) to produce inosine (I) in double-stranded (ds) RNA. This posttranscriptional RNA modification by adenosine deamination, known as A-to-I RNA editing, diversifies the transcriptome and modulates the innate immune interferon response. A-to-I editing is catalyzed by a family of enzymes, adenosine deaminases acting on dsRNA (ADARs). The roles of A-to-I editing are varied and include effects on mRNA translation, pre-mRNA splicing, and micro-RNA silencing. Suppression of dsRNA-triggered induction and action of interferon, the cornerstone of innate immunity, has emerged as a key function of ADAR1 editing of self (cellular) and nonself (viral) dsRNAs. A-to-I modification of RNA is essential for the normal regulation of cellular processes. Dysregulation of A-to-I editing by ADAR1 can have profound consequences, ranging from effects on cell growth and development to autoimmune disorders.
Keywords: 2'-5'-oligoadenylate synthetase; ADAR; RIG-I–like receptor (RLR); RNA deamination; RNA editing; adenosine deaminase acting on RNA; double-stranded RNA (dsRNA); innate immunity; interferon; protein kinase PKR.
© 2019 Samuel.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
Similar articles
-
Editing of Cellular Self-RNAs by Adenosine Deaminase ADAR1 Suppresses Innate Immune Stress Responses.J Biol Chem. 2016 Mar 18;291(12):6158-68. doi: 10.1074/jbc.M115.709014. Epub 2016 Jan 27. J Biol Chem. 2016. PMID: 26817845 Free PMC article.
-
An RNA editor, adenosine deaminase acting on double-stranded RNA (ADAR1).J Interferon Cytokine Res. 2014 Jun;34(6):437-46. doi: 10.1089/jir.2014.0001. J Interferon Cytokine Res. 2014. PMID: 24905200 Free PMC article. Review.
-
Adenosine Deaminases Acting on RNA (ADARs) and Viral Infections.Annu Rev Virol. 2021 Sep 29;8(1):239-264. doi: 10.1146/annurev-virology-091919-065320. Epub 2021 Apr 21. Annu Rev Virol. 2021. PMID: 33882257
-
ADAR RNA Modifications, the Epitranscriptome and Innate Immunity.Trends Biochem Sci. 2021 Sep;46(9):758-771. doi: 10.1016/j.tibs.2021.02.002. Epub 2021 Mar 15. Trends Biochem Sci. 2021. PMID: 33736931 Review.
-
Chimeric double-stranded RNA-specific adenosine deaminase ADAR1 proteins reveal functional selectivity of double-stranded RNA-binding domains from ADAR1 and protein kinase PKR.Proc Natl Acad Sci U S A. 2000 Nov 7;97(23):12541-6. doi: 10.1073/pnas.97.23.12541. Proc Natl Acad Sci U S A. 2000. PMID: 11070079 Free PMC article.
Cited by
-
mRNA Editing, Processing and Quality Control in Caenorhabditis elegans.Genetics. 2020 Jul;215(3):531-568. doi: 10.1534/genetics.119.301807. Genetics. 2020. PMID: 32632025 Free PMC article. Review.
-
Immune Sensing Mechanisms that Discriminate Self from Altered Self and Foreign Nucleic Acids.Immunity. 2020 Jul 14;53(1):54-77. doi: 10.1016/j.immuni.2020.06.014. Immunity. 2020. PMID: 32668228 Free PMC article. Review.
-
ALU non-B-DNA conformations, flipons, binary codes and evolution.R Soc Open Sci. 2020 Jun 3;7(6):200222. doi: 10.1098/rsos.200222. eCollection 2020 Jun. R Soc Open Sci. 2020. PMID: 32742689 Free PMC article. Review.
-
Deciphering the Biological Significance of ADAR1-Z-RNA Interactions.Int J Mol Sci. 2021 Oct 23;22(21):11435. doi: 10.3390/ijms222111435. Int J Mol Sci. 2021. PMID: 34768866 Free PMC article. Review.
-
An essential role of adenosine deaminase acting on RNA 1 in coeliac disease mucosa.Front Immunol. 2023 May 8;14:1175348. doi: 10.3389/fimmu.2023.1175348. eCollection 2023. Front Immunol. 2023. PMID: 37223095 Free PMC article.
References
-
- Wagner R. W., Smith J. E., Cooperman B. S., and Nishikura K. (1989) A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc. Natl. Acad. Sci. U.S.A. 86, 2647–2651 10.1073/pnas.86.8.2647 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

