Eukaryotic 5-methylcytosine (m⁵C) RNA Methyltransferases: Mechanisms, Cellular Functions, and Links to Disease
- PMID: 30704115
- PMCID: PMC6409601
- DOI: 10.3390/genes10020102
Eukaryotic 5-methylcytosine (m⁵C) RNA Methyltransferases: Mechanisms, Cellular Functions, and Links to Disease
Abstract
5-methylcytosine (m⁵C) is an abundant RNA modification that's presence is reported in a wide variety of RNA species, including cytoplasmic and mitochondrial ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs), as well as messenger RNAs (mRNAs), enhancer RNAs (eRNAs) and a number of non-coding RNAs. In eukaryotes, C5 methylation of RNA cytosines is catalyzed by enzymes of the NOL1/NOP2/SUN domain (NSUN) family, as well as the DNA methyltransferase homologue DNMT2. In recent years, substrate RNAs and modification target nucleotides for each of these methyltransferases have been identified, and structural and biochemical analyses have provided the first insights into how each of these enzymes achieves target specificity. Functional characterizations of these proteins and the modifications they install have revealed important roles in diverse aspects of both mitochondrial and nuclear gene expression. Importantly, this knowledge has enabled a better understanding of the molecular basis of a number of diseases caused by mutations in the genes encoding m⁵C methyltransferases or changes in the expression level of these enzymes.
Keywords: 5-methylcytosine; RNA methyltransferase; RNA modification; epitranscriptome; gene expression; messenger RNA (mRNA); mitochondria; ribosome; transfer RNA (tRNA).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


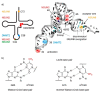
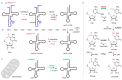
Similar articles
-
The RNA Methyltransferase NSUN2 and Its Potential Roles in Cancer.Cells. 2020 Jul 22;9(8):1758. doi: 10.3390/cells9081758. Cells. 2020. PMID: 32708015 Free PMC article. Review.
-
5-methylcytosine RNA methyltransferases and their potential roles in cancer.J Transl Med. 2022 May 13;20(1):214. doi: 10.1186/s12967-022-03427-2. J Transl Med. 2022. PMID: 35562754 Free PMC article. Review.
-
Eukaryotic rRNA Modification by Yeast 5-Methylcytosine-Methyltransferases and Human Proliferation-Associated Antigen p120.PLoS One. 2015 Jul 21;10(7):e0133321. doi: 10.1371/journal.pone.0133321. eCollection 2015. PLoS One. 2015. PMID: 26196125 Free PMC article.
-
The Roles of Host 5-Methylcytosine RNA Methyltransferases during Viral Infections.Int J Mol Sci. 2020 Oct 31;21(21):8176. doi: 10.3390/ijms21218176. Int J Mol Sci. 2020. PMID: 33142933 Free PMC article. Review.
-
Conservation of tRNA and rRNA 5-methylcytosine in the kingdom Plantae.BMC Plant Biol. 2015 Aug 14;15:199. doi: 10.1186/s12870-015-0580-8. BMC Plant Biol. 2015. PMID: 26268215 Free PMC article.
Cited by
-
Human NOP2/NSUN1 regulates ribosome biogenesis through non-catalytic complex formation with box C/D snoRNPs.Nucleic Acids Res. 2022 Oct 14;50(18):10695-10716. doi: 10.1093/nar/gkac817. Nucleic Acids Res. 2022. PMID: 36161484 Free PMC article.
-
An old friend with a new face: tRNA-derived small RNAs with big regulatory potential in cancer biology.Br J Cancer. 2023 May;128(9):1625-1635. doi: 10.1038/s41416-023-02191-4. Epub 2023 Feb 9. Br J Cancer. 2023. PMID: 36759729 Free PMC article. Review.
-
Prognostic Characteristics and Immune Effects of N6-Methyladenosine and 5-Methylcytosine-Related Regulatory Factors in Clear Cell Renal Cell Carcinoma.Front Genet. 2022 Apr 27;13:864383. doi: 10.3389/fgene.2022.864383. eCollection 2022. Front Genet. 2022. PMID: 35571068 Free PMC article.
-
Multi-omics analysis of expression and prognostic value of NSUN members in prostate cancer.Front Oncol. 2022 Aug 1;12:965571. doi: 10.3389/fonc.2022.965571. eCollection 2022. Front Oncol. 2022. PMID: 35978830 Free PMC article.
-
Change of Heart: the Epitranscriptome of Small Non-coding RNAs in Heart Failure.Curr Heart Fail Rep. 2022 Oct;19(5):255-266. doi: 10.1007/s11897-022-00561-2. Epub 2022 Jul 25. Curr Heart Fail Rep. 2022. PMID: 35876969 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

