Breakpoint mapping at nucleotide resolution in X-autosome balanced translocations associated with clinical phenotypes
- PMID: 30700833
- PMCID: PMC6461923
- DOI: 10.1038/s41431-019-0341-5
Breakpoint mapping at nucleotide resolution in X-autosome balanced translocations associated with clinical phenotypes
Abstract
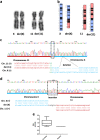
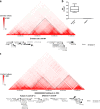
Precise breakpoint mapping of balanced chromosomal rearrangements is crucial to identify disease etiology. Ten female patients with X-autosome balanced translocations associated with phenotypic alterations were evaluated, by mapping and sequencing their breakpoints. The rearrangements' impact on the expression of disrupted genes, and inferred mechanisms of formation in each case were assessed. For four patients that presented one of the chromosomal breaks in heterochromatic and highly repetitive segments, we combined cytogenomic methods and short-read sequencing to characterize, at nucleotide resolution, breakpoints that occurred in reference genome gaps. Most of rearrangements were possibly formed by non-homologous end joining and have breakpoints at repeat elements. Seven genes were found to be disrupted in six patients. Six of the affected genes showed altered expression, and the functional impairment of three of them were considered pathogenic. One gene disruption was considered potentially pathogenic, and three had uncertain clinical significance. Four patients presented no gene disruptions, suggesting other pathogenic mechanisms. Four genes were considered potentially affected by position effect and the expression abrogation of one of them was confirmed. This study emphasizes the importance of breakpoint-junction characterization at nucleotide resolution in balanced rearrangements to reveal genetic mechanisms associated with the patients' phenotypes, mechanisms of formation that originated the rearrangements, and genomic nature of disrupted DNA sequences.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
Genetic mechanisms leading to primary amenorrhea in balanced X-autosome translocations.Fertil Steril. 2015 May;103(5):1289-96.e2. doi: 10.1016/j.fertnstert.2015.01.030. Epub 2015 Mar 4. Fertil Steril. 2015. PMID: 25747126
-
Breakpoint determination of X;autosome balanced translocations in four patients with premature ovarian failure.J Hum Genet. 2011 Feb;56(2):156-60. doi: 10.1038/jhg.2010.155. Epub 2010 Dec 9. J Hum Genet. 2011. PMID: 21150920
-
Accurate Breakpoint Mapping in Apparently Balanced Translocation Families with Discordant Phenotypes Using Whole Genome Mate-Pair Sequencing.PLoS One. 2017 Jan 10;12(1):e0169935. doi: 10.1371/journal.pone.0169935. eCollection 2017. PLoS One. 2017. PMID: 28072833 Free PMC article.
-
Historical and Clinical Perspectives on Chromosomal Translocations.Adv Exp Med Biol. 2018;1044:1-14. doi: 10.1007/978-981-13-0593-1_1. Adv Exp Med Biol. 2018. PMID: 29956287 Review.
-
Prenatal diagnosis of de novo X;autosome translocations.Clin Genet. 2004 May;65(5):423-8. doi: 10.1111/j.0009-9163.2004.00255.x. Clin Genet. 2004. PMID: 15099352 Review.
Cited by
-
Molecular Cytogenetics in the Era of Chromosomics and Cytogenomic Approaches.Front Genet. 2021 Oct 13;12:720507. doi: 10.3389/fgene.2021.720507. eCollection 2021. Front Genet. 2021. PMID: 34721522 Free PMC article. Review.
-
Coffin-Lowry syndrome in a girl with 46,XX,t(X;11)(p22;p15)dn: Identification of RPS6KA3 disruption by whole genome sequencing.Clin Case Rep. 2020 Apr 6;8(6):1076-1080. doi: 10.1002/ccr3.2826. eCollection 2020 Jun. Clin Case Rep. 2020. PMID: 32577269 Free PMC article.
-
The genetic cause of intellectual deficiency and/or congenital malformations in two parental reciprocal translocation carriers and implications for assisted reproduction.J Assist Reprod Genet. 2021 Jan;38(1):243-250. doi: 10.1007/s10815-020-01986-1. Epub 2020 Oct 22. J Assist Reprod Genet. 2021. PMID: 33094427 Free PMC article.
-
Allelic and dosage effects of NHS in X-linked cataract and Nance-Horan syndrome: a family study and literature review.Mol Cytogenet. 2021 Oct 7;14(1):48. doi: 10.1186/s13039-021-00566-x. Mol Cytogenet. 2021. PMID: 34620209 Free PMC article.
-
Mechanisms of structural chromosomal rearrangement formation.Mol Cytogenet. 2022 Jun 14;15(1):23. doi: 10.1186/s13039-022-00600-6. Mol Cytogenet. 2022. PMID: 35701783 Free PMC article. Review.
References
-
- Schluth-Bolard C, Labalme A, Cordier MP, Till M, Nadeau G, Tevissen H, et al. Breakpoint mapping by next generation sequencing reveals causative gene disruption in patients carrying apparently balanced chromosome rearrangements with intellectual deficiency and/or congenital malformations. J Med Genet. 2013;50:144–50. doi: 10.1136/jmedgenet-2012-101351. - DOI - PubMed
-
- Nilsson D, Pettersson M, Gustavsson P, Forster A, Hofmeister W, Wincent J, et al. Whole-Genome Sequencing of Cytogenetically Balanced Chromosome Translocations Identifies Potentially Pathological Gene Disruptions and Highlights the Importance of Microhomology in the Mechanism of Formation. Hum Mutat. 2017;38:180–92. doi: 10.1002/humu.23146. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

