hiPSCs in cardio-oncology: deciphering the genomics
- PMID: 30689737
- PMCID: PMC6452310
- DOI: 10.1093/cvr/cvz018
hiPSCs in cardio-oncology: deciphering the genomics
Abstract
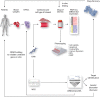
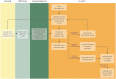
The genomic predisposition to oncology-drug-induced cardiovascular toxicity has been postulated for many decades. Only recently has it become possible to experimentally validate this hypothesis via the use of patient-specific human-induced pluripotent stem cells (hiPSCs) and suitably powered genome-wide association studies (GWAS). Identifying the individual single nucleotide polymorphisms (SNPs) responsible for the susceptibility to toxicity from a specific drug is a daunting task as this precludes the use of one of the most powerful tools in genomics: comparing phenotypes to close relatives, as these are highly unlikely to have been treated with the same drug. Great strides have been made through the use of candidate gene association studies (CGAS) and increasingly large GWAS studies, as well as in vivo whole-organism studies to further our mechanistic understanding of this toxicity. The hiPSC model is a powerful technology to build on this work and identify and validate causal variants in mechanistic pathways through directed genomic editing such as CRISPR. The causative variants identified through these studies can then be implemented clinically to identify those likely to experience cardiovascular toxicity and guide treatment options. Additionally, targets identified through hiPSC studies can inform future drug development. Through careful phenotypic characterization, identification of genomic variants that contribute to gene function and expression, and genomic editing to verify mechanistic pathways, hiPSC technology is a critical tool for drug discovery and the realization of precision medicine in cardio-oncology.
Keywords: Cardiotoxicity; Chemotherapy; Pharmacogenomics; Prediction; hiPSC.
Published on behalf of the European Society of Cardiology. All rights reserved. © The Author(s) 2019. For permissions, please email: journals.permissions@oup.com.
Figures
Similar articles
-
Personalized medicine in cardio-oncology: the role of induced pluripotent stem cell.Cardiovasc Res. 2019 Apr 15;115(5):949-959. doi: 10.1093/cvr/cvz024. Cardiovasc Res. 2019. PMID: 30768178 Free PMC article. Review.
-
Human In Vitro Models for Assessing the Genomic Basis of Chemotherapy-Induced Cardiovascular Toxicity.J Cardiovasc Transl Res. 2020 Jun;13(3):377-389. doi: 10.1007/s12265-020-09962-x. Epub 2020 Feb 20. J Cardiovasc Transl Res. 2020. PMID: 32078739 Free PMC article. Review.
-
Modeling Precision Cardio-Oncology: Using Human-Induced Pluripotent Stem Cells for Risk Stratification and Prevention.Curr Oncol Rep. 2021 May 3;23(7):77. doi: 10.1007/s11912-021-01066-2. Curr Oncol Rep. 2021. PMID: 33937943 Free PMC article. Review.
-
Validating the pharmacogenomics of chemotherapy-induced cardiotoxicity: What is missing?Pharmacol Ther. 2016 Dec;168:113-125. doi: 10.1016/j.pharmthera.2016.09.009. Epub 2016 Sep 5. Pharmacol Ther. 2016. PMID: 27609196 Free PMC article. Review.
-
Use of hiPSC to explicate genomic predisposition to anthracycline-induced cardiotoxicity.Pharmacogenomics. 2021 Jan;22(1):41-54. doi: 10.2217/pgs-2020-0104. Epub 2021 Jan 15. Pharmacogenomics. 2021. PMID: 33448871 Free PMC article. Review.
Cited by
-
Cancer Therapy-Related Cardiac Dysfunction: An Overview for the Clinician.Clin Med Insights Cardiol. 2019 Jul 29;13:1179546819866445. doi: 10.1177/1179546819866445. eCollection 2019. Clin Med Insights Cardiol. 2019. PMID: 31384135 Free PMC article. Review.
-
Cell Transdifferentiation and Reprogramming in Disease Modeling: Insights into the Neuronal and Cardiac Disease Models and Current Translational Strategies.Cells. 2021 Sep 27;10(10):2558. doi: 10.3390/cells10102558. Cells. 2021. PMID: 34685537 Free PMC article. Review.
-
Left Ventricular Systolic Function Has Strong Independent Genetic Background from Diastolic Function: A Classical Twin Study.Medicina (Kaunas). 2021 Sep 5;57(9):935. doi: 10.3390/medicina57090935. Medicina (Kaunas). 2021. PMID: 34577858 Free PMC article.
-
Proteomic Profiling Reveals Roles of Stress Response, Ca2+ Transient Dysregulation, and Novel Signaling Pathways in Alcohol-Induced Cardiotoxicity.Alcohol Clin Exp Res. 2020 Nov;44(11):2187-2199. doi: 10.1111/acer.14471. Epub 2020 Oct 16. Alcohol Clin Exp Res. 2020. PMID: 32981093 Free PMC article.
-
Biomarkers of Trastuzumab-Induced Cardiac Toxicity in HER2- Positive Breast Cancer Patient Population.Cancers (Basel). 2022 Jul 10;14(14):3353. doi: 10.3390/cancers14143353. Cancers (Basel). 2022. PMID: 35884413 Free PMC article.
References
-
- Lotrionte M, Biondi-Zoccai G, Abbate A, Lanzetta G, D'Ascenzo F, Malavasi V, Peruzzi M, Frati G, Palazzoni G.. Review and meta-analysis of incidence and clinical predictors of anthracycline cardiotoxicity. Am J Cardiol 2013;112:1980–1984. - PubMed
-
- Magdy T, Burridge PW.. The future role of pharmacogenomics in anticancer agent-induced cardiovascular toxicity. Pharmacogenomics 2018;19:79–82. - PubMed
-
- Colhoun HM, McKeigue PM, Davey Smith G.. Problems of reporting genetic associations with complex outcomes. Lancet 2003;361:865–872. - PubMed
-
- Hewitt JK. Editorial policy on candidate gene association and candidate gene-by-environment interaction studies of complex traits. Behav Genet 2012;42:1–2. - PubMed
-
- Pashos EE, Park Y, Wang X, Raghavan A, Yang W, Abbey D, Peters DT, Arbelaez J, Hernandez M, Kuperwasser N, Li W, Lian Z, Liu Y, Lv W, Lytle-Gabbin SL, Marchadier DH, Rogov P, Shi J, Slovik KJ, Stylianou IM, Wang L, Yan R, Zhang X, Kathiresan S, Duncan SA, Mikkelsen TS, Morrisey EE, Rader DJ, Brown CD, Musunuru K.. Large, diverse population cohorts of hiPSCs and derived hepatocyte-like cells reveal functional genetic variation at blood lipid-associated loci. Cell Stem Cell 2017;20:558–570 e510. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

