LANA oligomeric architecture is essential for KSHV nuclear body formation and viral genome maintenance during latency
- PMID: 30682185
- PMCID: PMC6364946
- DOI: 10.1371/journal.ppat.1007489
LANA oligomeric architecture is essential for KSHV nuclear body formation and viral genome maintenance during latency
Abstract
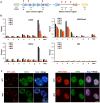
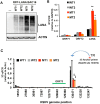
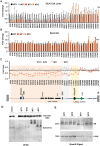
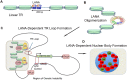
The molecular basis for the formation of functional, higher-ordered macro-molecular domains is not completely known. The Kaposi's Sarcoma-Associated Herpesvirus (KSHV) genome forms a super-molecular domain structure during latent infection that is strictly dependent on the DNA binding of the viral nuclear antigen LANA to the viral terminal repeats (TR). LANA is known to form oligomeric structures that have been implicated in viral episome maintenance. In this study, we show that the LANA oligomerization interface is required for the formation of higher-order nuclear bodies that partially colocalize with DAXX, EZH2, H3K27me3, and ORC2 but not with PML. These nuclear bodies assemble at the periphery of condensed cellular chromosomes during mitotic cell division. We demonstrate that the LANA oligomerization interface contributes to the cooperative DNA binding at the viral TR and the recruitment of ORC to the viral episome. Oligomerization mutants failed to auto-regulate LANA/ORF73 transcription, and this correlated with the loss of a chromosome conformational DNA-loop between the TR and LANA promoter. Viral genomes with LANA oligomerization mutants were subject to genome rearrangements including the loss of subgenomic DNA. Our data suggests that LANA oligomerization drives stable binding to the TR and formation of an epigenetically stable chromatin architecture resulting in higher-order LANA nuclear bodies important for viral genome integrity and long-term episome persistence.
Conflict of interest statement
Paul Lieberman is a founder of Vironika, LLC.
Figures
Similar articles
-
Kaposi's Sarcoma-Associated Herpesvirus LANA-Adjacent Regions with Distinct Functions in Episome Segregation or Maintenance.J Virol. 2019 Mar 5;93(6):e02158-18. doi: 10.1128/JVI.02158-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30626680 Free PMC article.
-
Identification of Kaposi's sarcoma-associated herpesvirus LANA regions important for episome segregation, replication, and persistence.J Virol. 2013 Nov;87(22):12270-83. doi: 10.1128/JVI.01243-13. Epub 2013 Sep 4. J Virol. 2013. PMID: 24006437 Free PMC article.
-
The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus supports latent DNA replication in dividing cells.J Virol. 2002 Nov;76(22):11677-87. doi: 10.1128/jvi.76.22.11677-11687.2002. J Virol. 2002. PMID: 12388727 Free PMC article.
-
KSHV LANA--the master regulator of KSHV latency.Viruses. 2014 Dec 11;6(12):4961-98. doi: 10.3390/v6124961. Viruses. 2014. PMID: 25514370 Free PMC article. Review.
-
Kaposi's Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen: Replicating and Shielding Viral DNA during Viral Persistence.J Virol. 2017 Jun 26;91(14):e01083-16. doi: 10.1128/JVI.01083-16. Print 2017 Jul 15. J Virol. 2017. PMID: 28446671 Free PMC article. Review.
Cited by
-
A Panel of Kaposi's Sarcoma-Associated Herpesvirus Mutants in the Polycistronic Kaposin Locus for Precise Analysis of Individual Protein Products.J Virol. 2022 Mar 9;96(5):e0156021. doi: 10.1128/JVI.01560-21. Epub 2021 Dec 22. J Virol. 2022. PMID: 34936820 Free PMC article.
-
Molecular mechanisms and cellular functions of liquid-liquid phase separation during antiviral immune responses.Front Immunol. 2023 May 10;14:1162211. doi: 10.3389/fimmu.2023.1162211. eCollection 2023. Front Immunol. 2023. PMID: 37251408 Free PMC article. Review.
-
Human Herpesvirus Sequencing in the Genomic Era: The Growing Ranks of the Herpetic Legion.Pathogens. 2019 Oct 12;8(4):186. doi: 10.3390/pathogens8040186. Pathogens. 2019. PMID: 31614759 Free PMC article. Review.
-
G-Quadruplexes in the Regulation of Viral Gene Expressions and Their Impacts on Controlling Infection.Pathogens. 2024 Jan 8;13(1):60. doi: 10.3390/pathogens13010060. Pathogens. 2024. PMID: 38251367 Free PMC article. Review.
-
Phase separation and DAXX redistribution contribute to LANA nuclear body and KSHV genome dynamics during latency and reactivation.PLoS Pathog. 2021 Jan 20;17(1):e1009231. doi: 10.1371/journal.ppat.1009231. eCollection 2021 Jan. PLoS Pathog. 2021. PMID: 33471863 Free PMC article.
References
-
- Lieberman PM, Hu J, Renne R. Maintenance and replication during latency In: Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, et al., editors. Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. Cambridge: 2007. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

