Population structure of human gut bacteria in a diverse cohort from rural Tanzania and Botswana
- PMID: 30665461
- PMCID: PMC6341659
- DOI: 10.1186/s13059-018-1616-9
Population structure of human gut bacteria in a diverse cohort from rural Tanzania and Botswana
Abstract
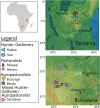
Background: Gut microbiota from individuals in rural, non-industrialized societies differ from those in individuals from industrialized societies. Here, we use 16S rRNA sequencing to survey the gut bacteria of seven non-industrialized populations from Tanzania and Botswana. These include populations practicing traditional hunter-gatherer, pastoralist, and agropastoralist subsistence lifestyles and a comparative urban cohort from the greater Philadelphia region.
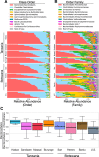
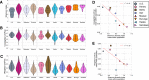
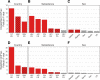
Results: We find that bacterial diversity per individual and within-population phylogenetic dissimilarity differs between Botswanan and Tanzanian populations, with Tanzania generally having higher diversity per individual and lower dissimilarity between individuals. Among subsistence groups, the gut bacteria of hunter-gatherers are phylogenetically distinct from both agropastoralists and pastoralists, but that of agropastoralists and pastoralists were not significantly different from each other. Nearly half of the Bantu-speaking agropastoralists from Botswana have gut bacteria that are very similar to the Philadelphian cohort. Based on imputed metagenomic content, US samples have a relative enrichment of genes found in pathways for degradation of several common industrial pollutants. Within two African populations, we find evidence that bacterial composition correlates with the genetic relatedness between individuals.
Conclusions: Across the cohort, similarity in bacterial presence/absence compositions between people increases with both geographic proximity and genetic relatedness, while abundance weighted bacterial composition varies more significantly with geographic proximity than with genetic relatedness.
Keywords: Adaptation; Agropastoralists; Diet; Genetics; Gut microbiome; Hunter-gatherers; Industrialization; Pastoralists; Rural populations; Sub-Saharan Africa.
Conflict of interest statement
Ethics approval and consent to participate
All experiments were approved by the University of Pennsylvania Institutional Review Board (IRB # 807981). Ethical approval and permits were also obtained from the following institutions prior to sample collection: The Commission for Science and Technology and National Institute for Medical Research in Dar es Salaam and the Ministry of Health in the Republic of Botswana. Written, informed consent was obtained from each participant.
All experimental methods were in accordance with Helsinki Declaration.
Consent for publication
All participants provided consent for publication of study results of the collected biomaterials paired with anonymized information on age, sex, and location.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


Similar articles
-
Lifestyle and the presence of helminths is associated with gut microbiome composition in Cameroonians.Genome Biol. 2020 May 25;21(1):122. doi: 10.1186/s13059-020-02020-4. Genome Biol. 2020. PMID: 32450885 Free PMC article.
-
Variation in Rural African Gut Microbiota Is Strongly Correlated with Colonization by Entamoeba and Subsistence.PLoS Genet. 2015 Nov 30;11(11):e1005658. doi: 10.1371/journal.pgen.1005658. eCollection 2015 Nov. PLoS Genet. 2015. PMID: 26619199 Free PMC article.
-
Subsistence strategies in traditional societies distinguish gut microbiomes.Nat Commun. 2015 Mar 25;6:6505. doi: 10.1038/ncomms7505. Nat Commun. 2015. PMID: 25807110 Free PMC article.
-
Meta-analysis of the human gut microbiome from urbanized and pre-agricultural populations.Environ Microbiol. 2017 Apr;19(4):1379-1390. doi: 10.1111/1462-2920.13692. Epub 2017 Feb 28. Environ Microbiol. 2017. PMID: 28198087 Review.
-
Geography, Ethnicity or Subsistence-Specific Variations in Human Microbiome Composition and Diversity.Front Microbiol. 2017 Jun 23;8:1162. doi: 10.3389/fmicb.2017.01162. eCollection 2017. Front Microbiol. 2017. PMID: 28690602 Free PMC article. Review.
Cited by
-
High abundance of butyrate-producing bacteria in the naso-oropharynx of SARS-CoV-2-infected persons in an African population: implications for low disease severity.BMC Infect Dis. 2024 Sep 20;24(1):1020. doi: 10.1186/s12879-024-09948-z. BMC Infect Dis. 2024. PMID: 39304808 Free PMC article.
-
The gut microbiome in konzo.Nat Commun. 2021 Sep 10;12(1):5371. doi: 10.1038/s41467-021-25694-1. Nat Commun. 2021. PMID: 34508085 Free PMC article.
-
The gut microbiome of Baka forager-horticulturalists from Cameroon is optimized for wild plant foods.iScience. 2024 Feb 10;27(3):109211. doi: 10.1016/j.isci.2024.109211. eCollection 2024 Mar 15. iScience. 2024. PMID: 38433907 Free PMC article.
-
Benchmark of 16S rRNA gene amplicon sequencing using Japanese gut microbiome data from the V1-V2 and V3-V4 primer sets.BMC Genomics. 2021 Jul 10;22(1):527. doi: 10.1186/s12864-021-07746-4. BMC Genomics. 2021. PMID: 34246242 Free PMC article.
-
Multicenter assessment of microbial community profiling using 16S rRNA gene sequencing and shotgun metagenomic sequencing.J Adv Res. 2020 Jul 21;26:111-121. doi: 10.1016/j.jare.2020.07.010. eCollection 2020 Nov. J Adv Res. 2020. PMID: 33133687 Free PMC article.
References
-
- Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Sears MR, et al. Infant gut microbiota and the hygiene hypothesis of allergic disease: impact of household pets and siblings on microbiota composition and diversity. Allergy Asthma Clin Immunol. 2013;9:15. doi: 10.1186/1710-1492-9-15. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

