Multicolour single-molecule tracking of mRNA interactions with RNP granules
- PMID: 30664789
- PMCID: PMC6375083
- DOI: 10.1038/s41556-018-0263-4
Multicolour single-molecule tracking of mRNA interactions with RNP granules
Abstract
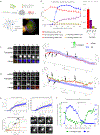
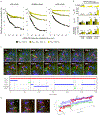
Ribonucleoprotein (RNP) granules are non-membrane-bound organelles that have critical roles in the stress response1,2, maternal messenger RNA storage3, synaptic plasticity4, tumour progression5,6 and neurodegeneration7-9. However, the dynamics of their mRNA components within and near the granule surface remain poorly characterized, particularly in the context and timing of mRNAs exiting translation. Herein, we used multicolour single-molecule tracking to quantify the precise timing and kinetics of single mRNAs as they exit translation and enter RNP granules during stress. We observed single mRNAs interacting with stress granules and P-bodies, with mRNAs moving bidirectionally between them. Although translating mRNAs only interact with RNP granules dynamically, non-translating mRNAs can form stable, and sometimes rigid, associations with RNP granules with stability increasing with both mRNA length and granule size. Live and fixed cell imaging demonstrated that mRNAs can extend beyond the protein surface of a stress granule, which may facilitate interactions between RNP granules. Thus, the recruitment of mRNPs to RNP granules involves dynamic, stable and extended interactions affected by translation status, mRNA length and granule size that collectively regulate RNP granule dynamics.
Figures


Comment in
-
Dynamics of mRNA entry into stress granules.Nat Cell Biol. 2019 Feb;21(2):116-117. doi: 10.1038/s41556-019-0278-5. Nat Cell Biol. 2019. PMID: 30664788 Free PMC article.
Similar articles
-
Eukaryotic stress granules: the ins and outs of translation.Mol Cell. 2009 Dec 25;36(6):932-41. doi: 10.1016/j.molcel.2009.11.020. Mol Cell. 2009. PMID: 20064460 Free PMC article. Review.
-
New insights into the regulation of RNP granule assembly in oocytes.Int Rev Cell Mol Biol. 2012;295:233-89. doi: 10.1016/B978-0-12-394306-4.00013-7. Int Rev Cell Mol Biol. 2012. PMID: 22449492 Free PMC article. Review.
-
Analyzing P-bodies and stress granules in Saccharomyces cerevisiae.Methods Enzymol. 2010;470:619-40. doi: 10.1016/S0076-6879(10)70025-2. Epub 2010 Mar 1. Methods Enzymol. 2010. PMID: 20946828
-
Principles and Properties of Stress Granules.Trends Cell Biol. 2016 Sep;26(9):668-679. doi: 10.1016/j.tcb.2016.05.004. Epub 2016 Jun 9. Trends Cell Biol. 2016. PMID: 27289443 Free PMC article. Review.
-
Direct observation of translational activation by a ribonucleoprotein granule.Nat Cell Biol. 2024 Aug;26(8):1322-1335. doi: 10.1038/s41556-024-01452-5. Epub 2024 Jul 4. Nat Cell Biol. 2024. PMID: 38965420 Free PMC article.
Cited by
-
mRNA Targeting, Transport and Local Translation in Eukaryotic Cells: From the Classical View to a Diversity of New Concepts.Mol Biol. 2021;55(4):507-537. doi: 10.1134/S0026893321030080. Epub 2021 May 30. Mol Biol. 2021. PMID: 34092811 Free PMC article.
-
Stress Granule Assembly Can Facilitate but Is Not Required for TDP-43 Cytoplasmic Aggregation.Biomolecules. 2020 Sep 25;10(10):1367. doi: 10.3390/biom10101367. Biomolecules. 2020. PMID: 32992901 Free PMC article.
-
Dynamics of mRNA entry into stress granules.Nat Cell Biol. 2019 Feb;21(2):116-117. doi: 10.1038/s41556-019-0278-5. Nat Cell Biol. 2019. PMID: 30664788 Free PMC article.
-
CNST is Characteristic of Leukemia Stem Cells and is Associated With Poor Prognosis in AML.Front Pharmacol. 2022 May 18;13:888243. doi: 10.3389/fphar.2022.888243. eCollection 2022. Front Pharmacol. 2022. PMID: 35662693 Free PMC article.
-
Optogenetic control of mRNA condensation reveals an intimate link between condensate material properties and functions.Nat Commun. 2024 Apr 15;15(1):3216. doi: 10.1038/s41467-024-47442-x. Nat Commun. 2024. PMID: 38622120 Free PMC article.
References
-
- El-Naggar AM & Sorensen PH Translational control of aberrant stress responses as a hallmark of cancer. J. Pathol 244, 650–666 (2018). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

