Family-based exome sequencing and case-control analysis implicate CEP41 as an ASD gene
- PMID: 30664616
- PMCID: PMC6341097
- DOI: 10.1038/s41398-018-0343-z
Family-based exome sequencing and case-control analysis implicate CEP41 as an ASD gene
Abstract
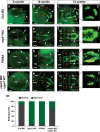
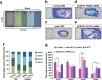
Autism Spectrum Disorder (ASD) is a complex neurodevelopmental disorder with a strong genetic component. Although next-generation sequencing (NGS) technologies have been successfully applied to gene identification in de novo ASD, the genetic architecture of familial ASD remains largely unexplored. Our approach, which leverages the high specificity and sensitivity of NGS technology, has focused on rare variants in familial autism. We used NGS exome sequencing in 26 families with distantly related affected individuals to identify genes with private gene disrupting and missense variants of interest (VOI). We found that the genes carrying VOIs were enriched for biological processes related to cell projection organization and neuron development, which is consistent with the neurodevelopmental hypothesis of ASD. For a subset of genes carrying VOIs, we then used targeted NGS sequencing and gene-based variant burden case-control analysis to test for association with ASD. Missense variants in one gene, CEP41, associated significantly with ASD (p = 6.185e-05). Homozygous gene-disrupting variants in CEP41 were initially found to be responsible for recessive Joubert syndrome. Using a zebrafish model, we evaluated the mechanism by which the CEP41 variants might contribute to ASD. We found that CEP41 missense variants affect development of the axonal tract, cranial neural crest migration and social behavior phenotype. Our work demonstrates the involvement of CEP41 heterozygous missense variants in ASD and that biological processes involved in cell projection organization and neuron development are enriched in ASD families we have studied.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Whole Exome Sequencing Identifies Novel De Novo Variants Interacting with Six Gene Networks in Autism Spectrum Disorder.Genes (Basel). 2020 Dec 22;12(1):1. doi: 10.3390/genes12010001. Genes (Basel). 2020. PMID: 33374967 Free PMC article.
-
Leveraging blood serotonin as an endophenotype to identify de novo and rare variants involved in autism.Mol Autism. 2017 Mar 21;8:14. doi: 10.1186/s13229-017-0130-3. eCollection 2017. Mol Autism. 2017. PMID: 28344757 Free PMC article.
-
Sequencing of sporadic Attention-Deficit Hyperactivity Disorder (ADHD) identifies novel and potentially pathogenic de novo variants and excludes overlap with genes associated with autism spectrum disorder.Am J Med Genet B Neuropsychiatr Genet. 2017 Jun;174(4):381-389. doi: 10.1002/ajmg.b.32527. Epub 2017 Mar 22. Am J Med Genet B Neuropsychiatr Genet. 2017. PMID: 28332277 Free PMC article.
-
Next-Generation Sequencing in Autism Spectrum Disorder.Cold Spring Harb Perspect Med. 2019 Aug 1;9(8):a026872. doi: 10.1101/cshperspect.a026872. Cold Spring Harb Perspect Med. 2019. PMID: 30420340 Free PMC article. Review.
-
Genetic architecture of autism spectrum disorder: Lessons from large-scale genomic studies.Neurosci Biobehav Rev. 2021 Sep;128:244-257. doi: 10.1016/j.neubiorev.2021.06.028. Epub 2021 Jun 21. Neurosci Biobehav Rev. 2021. PMID: 34166716 Review.
Cited by
-
Genotype-phenotype correlations and novel molecular insights into the DHX30-associated neurodevelopmental disorders.Genome Med. 2021 May 21;13(1):90. doi: 10.1186/s13073-021-00900-3. Genome Med. 2021. PMID: 34020708 Free PMC article.
-
Regulatory responses to assisted reproductive technology: a comparative analysis of Spain and Israel.J Assist Reprod Genet. 2019 Aug;36(8):1665-1681. doi: 10.1007/s10815-019-01525-7. Epub 2019 Jul 25. J Assist Reprod Genet. 2019. PMID: 31346936 Free PMC article.
-
Autistic Behavior as Novel Clinical Finding in OFD1 Syndrome.Genes (Basel). 2023 Jan 27;14(2):327. doi: 10.3390/genes14020327. Genes (Basel). 2023. PMID: 36833254 Free PMC article.
-
Molecular Dysregulation in Autism Spectrum Disorder.J Pers Med. 2021 Aug 27;11(9):848. doi: 10.3390/jpm11090848. J Pers Med. 2021. PMID: 34575625 Free PMC article. Review.
-
Insights Gained From Zebrafish Models for the Ciliopathy Joubert Syndrome.Front Genet. 2022 Jun 30;13:939527. doi: 10.3389/fgene.2022.939527. eCollection 2022. Front Genet. 2022. PMID: 35846153 Free PMC article. Review.
References
-
- Association AP. Diagnostic and Statistical Manual of Mental Disorders: Dsm-5. Washington, DC (American Psychiatric Publication Incorporated, 2013).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

