Targeted Endoplasmic Reticulum Localization of Storage Protein mRNAs Requires the RNA-Binding Protein RBP-L
- PMID: 30659066
- PMCID: PMC6393789
- DOI: 10.1104/pp.18.01434
Targeted Endoplasmic Reticulum Localization of Storage Protein mRNAs Requires the RNA-Binding Protein RBP-L
Abstract
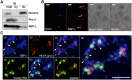
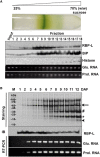
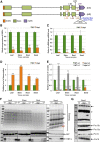
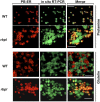
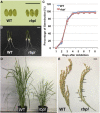
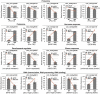
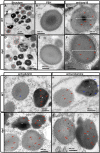
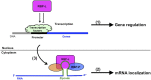
The transport and targeting of glutelin and prolamine mRNAs to distinct subdomains of the cortical endoplasmic reticulum is a model for mRNA localization in plants. This process requires a number of RNA-binding proteins (RBPs) that recognize and bind to mRNA cis-localization (zipcode) elements to form messenger ribonucleoprotein complexes, which then transport the RNAs to their destination sites at the cortical endoplasmic reticulum. Here, we present evidence that the rice (Oryza sativa) RNA-binding protein, RBP-L, like its interacting RBP-P partner, specifically binds to glutelin and prolamine zipcode RNA sequences and is required for proper mRNA localization in rice endosperm cells. A transfer DNA insertion in the 3' untranslated region resulted in reduced expression of the RBP-L gene to 10% to 25% of that in the wild-type. Reduced amounts of RBP-L caused partial mislocalization of glutelin and prolamine RNAs and conferred other general growth defects, including dwarfism, late flowering, and smaller seeds. Transcriptome analysis showed that RBP-L knockdown greatly affected the expression of prolamine family genes and several classes of transcription factors. Collectively, these results indicate that RBP-L, like RBP-P, is a key RBP involved in mRNA localization in rice endosperm cells. Moreover, distinct from RBP-P, RBP-L exhibits additional regulatory roles in development, either directly through its binding to corresponding RNAs or indirectly through its effect on transcription factors.
© 2019 American Society of Plant Biologists. All Rights Reserved.
Figures



Similar articles
-
Zipcode RNA-Binding Proteins and Membrane Trafficking Proteins Cooperate to Transport Glutelin mRNAs in Rice Endosperm.Plant Cell. 2020 Aug;32(8):2566-2581. doi: 10.1105/tpc.20.00111. Epub 2020 May 29. Plant Cell. 2020. PMID: 32471860 Free PMC article.
-
RNA-Binding Protein RBP-P Is Required for Glutelin and Prolamine mRNA Localization in Rice Endosperm Cells.Plant Cell. 2018 Oct;30(10):2529-2552. doi: 10.1105/tpc.18.00321. Epub 2018 Sep 6. Plant Cell. 2018. PMID: 30190374 Free PMC article.
-
Multiple RNA binding protein complexes interact with the rice prolamine RNA cis-localization zipcode sequences.Plant Physiol. 2014 Mar;164(3):1271-82. doi: 10.1104/pp.113.234187. Epub 2014 Jan 31. Plant Physiol. 2014. PMID: 24488967 Free PMC article.
-
The rice storage protein mRNAs as a model system for RNA localization in higher plants.Plant Sci. 2019 Jul;284:203-211. doi: 10.1016/j.plantsci.2019.04.014. Epub 2019 Apr 17. Plant Sci. 2019. PMID: 31084873 Review.
-
The role of mRNA and protein sorting in seed storage protein synthesis, transport, and deposition.Biochem Cell Biol. 2005 Dec;83(6):728-37. doi: 10.1139/o05-156. Biochem Cell Biol. 2005. PMID: 16333324 Review.
Cited by
-
Zipcode RNA-Binding Proteins and Membrane Trafficking Proteins Cooperate to Transport Glutelin mRNAs in Rice Endosperm.Plant Cell. 2020 Aug;32(8):2566-2581. doi: 10.1105/tpc.20.00111. Epub 2020 May 29. Plant Cell. 2020. PMID: 32471860 Free PMC article.
-
DGW1, encoding an hnRNP-like RNA binding protein, positively regulates grain size and weight by interacting with GW6 mRNA.Plant Biotechnol J. 2024 Feb;22(2):512-526. doi: 10.1111/pbi.14202. Epub 2023 Oct 20. Plant Biotechnol J. 2024. PMID: 37862261 Free PMC article.
-
Mechanisms and consequences of subcellular RNA localization across diverse cell types.Traffic. 2020 Jun;21(6):404-418. doi: 10.1111/tra.12730. Epub 2020 Apr 29. Traffic. 2020. PMID: 32291836 Free PMC article. Review.
-
Genes and Their Molecular Functions Determining Seed Structure, Components, and Quality of Rice.Rice (N Y). 2022 Mar 18;15(1):18. doi: 10.1186/s12284-022-00562-8. Rice (N Y). 2022. PMID: 35303197 Free PMC article. Review.
-
mRNA Localization in Plant Cells.Plant Physiol. 2020 Jan;182(1):97-109. doi: 10.1104/pp.19.00972. Epub 2019 Oct 14. Plant Physiol. 2020. PMID: 31611420 Free PMC article. Review.
References
-
- Baron KN, Schroeder DF, Stasolla C (2014) GEm-Related 5 (GER5), an ABA and stress-responsive GRAM domain protein regulating seed development and inflorescence architecture. Plant Sci 223: 153–166 - PubMed
-
- Belkhadir Y, Jaillais Y (2015) The molecular circuitry of brassinosteroid signaling. New Phytol 206: 522–540 - PubMed
-
- Bilenoglu O, Basak AN, Russell JE (2002) A 3'UTR mutation affects beta-globin expression without altering the stability of its fully processed mRNA. Br J Haematol 119: 1106–1114 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

