OLGA: fast computation of generation probabilities of B- and T-cell receptor amino acid sequences and motifs
- PMID: 30657870
- PMCID: PMC6735909
- DOI: 10.1093/bioinformatics/btz035
OLGA: fast computation of generation probabilities of B- and T-cell receptor amino acid sequences and motifs
Abstract
Motivation: High-throughput sequencing of large immune repertoires has enabled the development of methods to predict the probability of generation by V(D)J recombination of T- and B-cell receptors of any specific nucleotide sequence. These generation probabilities are very non-homogeneous, ranging over 20 orders of magnitude in real repertoires. Since the function of a receptor really depends on its protein sequence, it is important to be able to predict this probability of generation at the amino acid level. However, brute-force summation over all the nucleotide sequences with the correct amino acid translation is computationally intractable. The purpose of this paper is to present a solution to this problem.
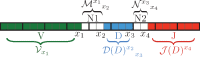
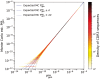
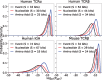
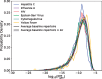
Results: We use dynamic programming to construct an efficient and flexible algorithm, called OLGA (Optimized Likelihood estimate of immunoGlobulin Amino-acid sequences), for calculating the probability of generating a given CDR3 amino acid sequence or motif, with or without V/J restriction, as a result of V(D)J recombination in B or T cells. We apply it to databases of epitope-specific T-cell receptors to evaluate the probability that a typical human subject will possess T cells responsive to specific disease-associated epitopes. The model prediction shows an excellent agreement with published data. We suggest that OLGA may be a useful tool to guide vaccine design.
Availability and implementation: Source code is available at https://github.com/zsethna/OLGA.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2019. Published by Oxford University Press.
Figures

Similar articles
-
repgenHMM: a dynamic programming tool to infer the rules of immune receptor generation from sequence data.Bioinformatics. 2016 Jul 1;32(13):1943-51. doi: 10.1093/bioinformatics/btw112. Epub 2016 Feb 26. Bioinformatics. 2016. PMID: 27153709 Free PMC article.
-
SOS: online probability estimation and generation of T-and B-cell receptors.Bioinformatics. 2020 Aug 15;36(16):4510-4512. doi: 10.1093/bioinformatics/btaa574. Bioinformatics. 2020. PMID: 32814974
-
IMGT(®) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS.Methods Mol Biol. 2012;882:569-604. doi: 10.1007/978-1-61779-842-9_32. Methods Mol Biol. 2012. PMID: 22665256
-
Revealing factors determining immunodominant responses against dominant epitopes.Immunogenetics. 2020 Feb;72(1-2):109-118. doi: 10.1007/s00251-019-01134-9. Epub 2019 Dec 6. Immunogenetics. 2020. PMID: 31811313 Free PMC article. Review.
-
RAG Chromatin Scanning During V(D)J Recombination and Chromatin Loop Extrusion are Related Processes.Adv Immunol. 2018;139:93-135. doi: 10.1016/bs.ai.2018.07.001. Epub 2018 Aug 27. Adv Immunol. 2018. PMID: 30249335 Review.
Cited by
-
Comprehensive Analysis of CDR3 Sequences in Gluten-Specific T-Cell Receptors Reveals a Dominant R-Motif and Several New Minor Motifs.Front Immunol. 2021 Apr 13;12:639672. doi: 10.3389/fimmu.2021.639672. eCollection 2021. Front Immunol. 2021. PMID: 33927715 Free PMC article.
-
Next-Generation Sequencing of T and B Cell Receptor Repertoires from COVID-19 Patients Showed Signatures Associated with Severity of Disease.Immunity. 2020 Aug 18;53(2):442-455.e4. doi: 10.1016/j.immuni.2020.06.024. Epub 2020 Jun 30. Immunity. 2020. PMID: 32668194 Free PMC article.
-
Echidna: integrated simulations of single-cell immune receptor repertoires and transcriptomes.Bioinform Adv. 2022 Sep 2;2(1):vbac062. doi: 10.1093/bioadv/vbac062. eCollection 2022. Bioinform Adv. 2022. PMID: 36699357 Free PMC article.
-
Population variability in the generation and selection of T-cell repertoires.PLoS Comput Biol. 2020 Dec 9;16(12):e1008394. doi: 10.1371/journal.pcbi.1008394. eCollection 2020 Dec. PLoS Comput Biol. 2020. PMID: 33296360 Free PMC article.
-
simAIRR: simulation of adaptive immune repertoires with realistic receptor sequence sharing for benchmarking of immune state prediction methods.Gigascience. 2022 Dec 28;12:giad074. doi: 10.1093/gigascience/giad074. Epub 2023 Oct 17. Gigascience. 2022. PMID: 37848619 Free PMC article.
References
-
- Becattini S. et al. (2015) Functional heterogeneity of human memory cd4+ t cell clones primed by pathogens or vaccines. Science, 347, 400–406. - PubMed
-
- Dupic T. et al. (2018) Genesis of the T-cell receptor. arXiv: 1806.11030.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

