Identification of potential genes and pathways for response prediction of neoadjuvant chemoradiotherapy in patients with rectal cancer by systemic biological analysis
- PMID: 30655792
- PMCID: PMC6313202
- DOI: 10.3892/ol.2018.9598
Identification of potential genes and pathways for response prediction of neoadjuvant chemoradiotherapy in patients with rectal cancer by systemic biological analysis
Abstract
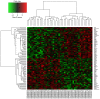
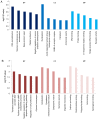
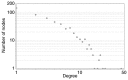
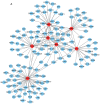
Currently, neoadjuvant chemoradiotherapy (CRT) followed by radical surgery is the standard of care for locally advanced rectal cancer. However, to the best of our knowledge, there are no effective biomarkers for predicting patients who may benefit from neoadjuvant treatment. The aim of the current study was to screen potential crucial genes and pathways associated with the response to CRT in rectal cancer, and provide valid biological information to assist further investigation of CRT optimization. In the current study, differentially expressed (DE) genes were identified from the tumor samples of responders and non-responders to neoadjuvant CRT in the GSE35452 gene expression profile. Seven hub genes and one significant module were identified from the protein-protein interaction (PPI) network. Functional enrichment analysis of all the DE genes and the hub genes, retrieved from PPI network analysis, revealed their associations with CRT response. Genes were identified that may be used to discriminate patients who would or would not clinically benefit from neoadjuvant CRT. Several important pathways enriched by the DE genes, hub genes and selected module were identified, and revealed to be closely associated with radiation response, including excision repair, homologous recombination, Ras signaling pathway, the forkhead box O signaling pathway, focal adhesion and the Wnt signaling pathway. In conclusion, the current study demonstrated that the identified gene signatures and pathways may be used as molecular biomarkers for predicting CRT response. Furthermore, combinations of these biomarkers may be helpful for optimizing CRT treatment and promoting understanding of the molecular basis of response differences; this needs to be confirmed by further experiments.
Keywords: differentially expressed genes; enrichment analysis; pathways; rectal cancer.
Figures

Similar articles
-
[Identification of gene biomarkers to predict responses to neoadjuvant chemoradiotherapy in patients with rectal cancer and pathways enrichment analysis].Zhonghua Wei Chang Wai Ke Za Zhi. 2019 Dec 25;22(12):1183-1187. doi: 10.3760/cma.j.issn.1671-0274.2019.12.015. Zhonghua Wei Chang Wai Ke Za Zhi. 2019. PMID: 31874536 Chinese.
-
Effect of Akt activation and experimental pharmacological inhibition on responses to neoadjuvant chemoradiotherapy in rectal cancer.Br J Surg. 2018 Jan;105(2):e192-e203. doi: 10.1002/bjs.10695. Br J Surg. 2018. PMID: 29341150
-
Predicting response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer with serum biomarkers.Ann R Coll Surg Engl. 2017 May;99(5):373-377. doi: 10.1308/rcsann.2017.0030. Ann R Coll Surg Engl. 2017. PMID: 28462648 Free PMC article.
-
Clinical utility of pretreatment prediction of chemoradiotherapy response in rectal cancer: a review.EPMA J. 2017 Mar 3;8(1):61-67. doi: 10.1007/s13167-017-0082-x. eCollection 2017 Mar. EPMA J. 2017. PMID: 28620444 Free PMC article. Review.
-
Complete response after neoadjuvant therapy in rectal cancer: to operate or not to operate?Dig Dis. 2012;30 Suppl 2:109-17. doi: 10.1159/000342039. Epub 2012 Nov 23. Dig Dis. 2012. PMID: 23207942 Review.
Cited by
-
Identification of Key Gene Targets for Sensitizing Colorectal Cancer to Chemoradiation: an Integrative Network Analysis on Multiple Transcriptomics Data.J Gastrointest Cancer. 2022 Sep;53(3):649-668. doi: 10.1007/s12029-021-00690-2. Epub 2021 Aug 25. J Gastrointest Cancer. 2022. PMID: 34432208
References
-
- Erlandsson J, Holm T, Pettersson D, Berglund Å, Cedermark B, Radu C, Johansson H, Machado M, Hjern F, Hallböök O, et al. Optimal fractionation of preoperative radiotherapy and timing to surgery for rectal cancer (Stockholm III): A multicentre, randomised, non-blinded, phase 3, non-inferiority trial. Lancet Oncol. 2017;18:336–346. doi: 10.1016/S1470-2045(17)30086-4. - DOI - PubMed
-
- Kim KH, Shin SJ, Cho MS, Ahn JB, Jung M, Kim TI, Park YS, Kim H, Kim NK, Koom WS. A phase II study of preoperative mFOLFOX6 with short-course radiotherapy in patients with locally advanced rectal cancer and liver-only metastasis. Radiother Oncol. 2016;118:369–374. doi: 10.1016/j.radonc.2015.11.029. - DOI - PubMed
-
- Burbach JP, den Harder AM, Intven M, van Vulpen M, Verkooijen HM, Reerink O. Impact of radiotherapy boost on pathological complete response in patients with locally advanced rectal cancer: A systematic review and meta-analysis. Radiother Oncol. 2014;113:1–9. doi: 10.1016/j.radonc.2014.08.035. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
