The Polar Region of the HIV-1 Envelope Protein Determines Viral Fusion and Infectivity by Stabilizing the gp120-gp41 Association
- PMID: 30651369
- PMCID: PMC6430531
- DOI: 10.1128/JVI.02128-18
The Polar Region of the HIV-1 Envelope Protein Determines Viral Fusion and Infectivity by Stabilizing the gp120-gp41 Association
Abstract
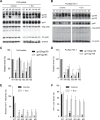
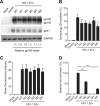
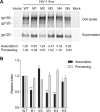
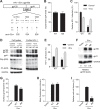
HIV-1 enters cells through binding between viral envelope glycoprotein (Env) and cellular receptors to initiate virus and cell fusion. HIV-1 Env precursor (gp160) is cleaved into two units noncovalently bound to form a trimer on virions, including a surface unit (gp120) and a transmembrane unit (gp41) responsible for virus binding and membrane fusion, respectively. The polar region (PR) at the N terminus of gp41 comprises 17 residues, including 7 polar amino acids. Previous studies suggested that the PR contributes to HIV-1 membrane fusion and infectivity; however, the precise role of the PR in Env-mediated viral entry and the underlying mechanisms remain unknown. Here, we show that the PR is critical for HIV-1 fusion and infectivity by stabilizing Env trimers. Through analyzing the PR sequences of 57,645 HIV-1 isolates, we performed targeted mutagenesis and functional studies of three highly conserved polar residues in the PR (S532P, T534A, and T536A) which have not been characterized previously. We found that single or combined mutations of these three residues abolished or significantly decreased HIV-1 infectivity without affecting viral production. These PR mutations abolished or significantly reduced HIV-1 fusion with target cells and also Env-mediated cell-cell fusion. Three PR mutations containing S532P substantially reduced gp120 and gp41 association, Env trimer stability, and increased gp120 shedding. Furthermore, S532A mutation significantly reduced HIV-1 infectivity and fusogenicity but not Env expression and cleavage. Our findings suggest that the PR of gp41, particularly the key residue S532, is structurally essential for maintaining HIV-1 Env trimer, viral fusogenicity, and infectivity.IMPORTANCE Although extensive studies of the transmembrane unit (gp41) of HIV-1 Env have led to a fusion inhibitor clinically used to block viral entry, the functions of different domains of gp41 in HIV-1 fusion and infectivity are not fully elucidated. The polar region (PR) of gp41 has been proposed to participate in HIV-1 membrane fusion in biochemical analyses, but its role in viral entry and infectivity remain unclear. In our effort to characterize three nucleotide mutations of an HIV-1 RNA element that partially overlaps the PR coding sequence, we identified a novel function of the PR that determines viral fusion and infectivity. We further demonstrated the structural and functional impact of six PR mutations on HIV-1 Env stability, viral fusion, and infectivity. Our findings reveal the previously unappreciated function of the PR and the underlying mechanisms, highlighting the important role of the PR in regulating HIV-1 fusion and infectivity.
Keywords: HIV-1; entry; envelope glycoprotein; fusion; gp160; gp41; infectivity; polar region; trimer.
Copyright © 2019 American Society for Microbiology.
Figures



Similar articles
-
Reverted HIV-1 Mutants in CD4+ T-Cells Reveal Critical Residues in the Polar Region of Viral Envelope Glycoprotein.Microbiol Spectr. 2021 Dec 22;9(3):e0165321. doi: 10.1128/spectrum.01653-21. Epub 2021 Dec 22. Microbiol Spectr. 2021. PMID: 34935422 Free PMC article.
-
HIV-1 gp41 Residues Modulate CD4-Induced Conformational Changes in the Envelope Glycoprotein and Evolution of a Relaxed Conformation of gp120.J Virol. 2018 Jul 31;92(16):e00583-18. doi: 10.1128/JVI.00583-18. Print 2018 Aug 15. J Virol. 2018. PMID: 29875245 Free PMC article.
-
Mutations That Increase the Stability of the Postfusion gp41 Conformation of the HIV-1 Envelope Glycoprotein Are Selected by both an X4 and R5 HIV-1 Virus To Escape Fusion Inhibitors Corresponding to Heptad Repeat 1 of gp41, but the gp120 Adaptive Mutations Differ between the Two Viruses.J Virol. 2019 May 15;93(11):e00142-19. doi: 10.1128/JVI.00142-19. Print 2019 Jun 1. J Virol. 2019. PMID: 30894471 Free PMC article.
-
HIV-1 gp41: mediator of fusion and target for inhibition.AIDS Rev. 2003 Oct-Dec;5(4):214-21. AIDS Rev. 2003. PMID: 15012000 Review.
-
HIV-1 replication.Somat Cell Mol Genet. 2001 Nov;26(1-6):13-33. doi: 10.1023/a:1021070512287. Somat Cell Mol Genet. 2001. PMID: 12465460 Review.
Cited by
-
Conserved Residue Asn-145 in the C-Terminal Heptad Repeat Region of HIV-1 gp41 is Critical for Viral Fusion and Regulates the Antiviral Activity of Fusion Inhibitors.Viruses. 2019 Jul 3;11(7):609. doi: 10.3390/v11070609. Viruses. 2019. PMID: 31277353 Free PMC article.
-
Functional Impacts of Epitranscriptomic m6A Modification on HIV-1 Infection.Viruses. 2024 Jan 16;16(1):127. doi: 10.3390/v16010127. Viruses. 2024. PMID: 38257827 Free PMC article. Review.
-
Identification of human immunodeficiency virus -1 E protein-targeting lead compounds by pharmacophore based screening.Saudi Med J. 2022 Dec;43(12):1324-1332. doi: 10.15537/smj.2022.43.12.20220599. Saudi Med J. 2022. PMID: 36517066 Free PMC article.
-
Alterations in gp120 glycans or the gp41 fusion peptide-proximal region modulate the stability of the human immunodeficiency virus (HIV-1) envelope glycoprotein pretriggered conformation.J Virol. 2023 Sep 28;97(9):e0059223. doi: 10.1128/jvi.00592-23. Epub 2023 Sep 11. J Virol. 2023. PMID: 37696048 Free PMC article.
-
HIV-1 envelope facilitates the development of protease inhibitor resistance through acquiring mutations associated with viral entry and immune escape.Front Microbiol. 2024 Apr 18;15:1388729. doi: 10.3389/fmicb.2024.1388729. eCollection 2024. Front Microbiol. 2024. PMID: 38699474 Free PMC article.
References
-
- Peisajovich SG, Epand RF, Pritsker M, Shai Y, Epand RM. 2000. The polar region consecutive to the HIV fusion peptide participates in membrane fusion. Biochemistry 39:1826–1833. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

