Reduction of Kaposi's Sarcoma-Associated Herpesvirus Latency Using CRISPR-Cas9 To Edit the Latency-Associated Nuclear Antigen Gene
- PMID: 30651362
- PMCID: PMC6430552
- DOI: 10.1128/JVI.02183-18
Reduction of Kaposi's Sarcoma-Associated Herpesvirus Latency Using CRISPR-Cas9 To Edit the Latency-Associated Nuclear Antigen Gene
Abstract
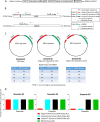
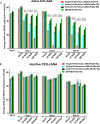
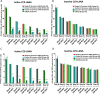
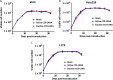
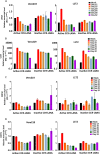
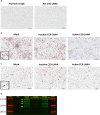
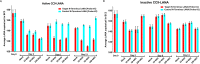
Kaposi's sarcoma-associated herpesvirus (KSHV) is the etiologic agent of Kaposi's sarcoma (KS), an AIDS-defining cancer in HIV-1-infected individuals or immune-suppressed transplant patients. The prevalence for both KSHV and KS are highest in sub-Saharan Africa where HIV-1 infection is also epidemic. There is no effective treatment for advanced KS; therefore, the survival rate is low. Similar to other herpesviruses, KSHV's ability to establish latent infection in the host presents a major challenge to KS treatment or prevention. Strategies to reduce KSHV episomal persistence in latently infected cells might lead to approaches to prevent KS development. The CRISPR-Cas9 system is a gene editing technique that has been used to specifically manipulate the HIV-1 genome but also Epstein-Barr virus (EBV) which, similar to KSHV, belongs to the Gammaherpesvirus family. Among KSHV gene products, the latency-associated nuclear antigen (LANA) is absolutely required in the maintenance, replication, and segregation of KSHV episomes during mitosis, which makes LANA an ideal target for CRISPR-Cas9 editing. In this study, we designed a replication-incompetent adenovirus type 5 to deliver a LANA-specific Cas9 system (Ad-CC9-LANA) into various KSHV latent target cells. We showed that KSHV latently infected epithelial and endothelial cells transduced with Ad-CC9-LANA underwent significant reductions in the KSHV episome burden, LANA RNA and protein expression over time, but this effect is less profound in BC3 cells due to the low infection efficiency of adenovirus type 5 for B cells. The use of an adenovirus vector might confer potential in vivo applications of LANA-specific Cas9 against KSHV infection and KS.IMPORTANCE The ability for Kaposi's sarcoma-associated herpesvirus (KSHV), the causative agent of Kaposi's sarcoma (KS), to establish and maintain latency has been a major challenge to clearing infection and preventing KS development. This is the first study to demonstrate the feasibility of using a KSHV LANA-targeted CRISPR-Cas9 and adenoviral delivery system to disrupt KSHV latency in infected epithelial and endothelial cell lines. Our system significantly reduced the KSHV episomal burden over time. Given the safety record of adenovirus as vaccine or delivery vectors, this approach to limit KSHV latency may also represent a viable strategy against other tumorigenic viruses and may have potential benefits in developing countries where the viral cancer burden is high.
Keywords: CRISPR-Cas9; Kaposi’s sarcoma-associated herpesvirus; LANA; latency.
Copyright © 2019 American Society for Microbiology.
Figures
Similar articles
-
Full-Length Isoforms of Kaposi's Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen Accumulate in the Cytoplasm of Cells Undergoing the Lytic Cycle of Replication.J Virol. 2017 Nov 30;91(24):e01532-17. doi: 10.1128/JVI.01532-17. Print 2017 Dec 15. J Virol. 2017. PMID: 28978712 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus LANA-Adjacent Regions with Distinct Functions in Episome Segregation or Maintenance.J Virol. 2019 Mar 5;93(6):e02158-18. doi: 10.1128/JVI.02158-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30626680 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus-encoded LANA contributes to viral latent replication by activating phosphorylation of survivin.J Virol. 2014 Apr;88(8):4204-17. doi: 10.1128/JVI.03855-13. Epub 2014 Jan 29. J Virol. 2014. PMID: 24478433 Free PMC article.
-
KSHV Genome Replication and Maintenance in Latency.Adv Exp Med Biol. 2018;1045:299-320. doi: 10.1007/978-981-10-7230-7_14. Adv Exp Med Biol. 2018. PMID: 29896673 Review.
-
Viral latent proteins as targets for Kaposi's sarcoma and Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) induced lymphoma.Curr Drug Targets Infect Disord. 2003 Jun;3(2):129-35. doi: 10.2174/1568005033481150. Curr Drug Targets Infect Disord. 2003. PMID: 12769790 Review.
Cited by
-
Targeting the Kaposi's sarcoma-associated herpesvirus genome with the CRISPR-Cas9 platform in latently infected cells.Virol J. 2021 Mar 17;18(1):56. doi: 10.1186/s12985-021-01527-x. Virol J. 2021. PMID: 33731154 Free PMC article.
-
Control of Viral Latency by Episome Maintenance Proteins.Trends Microbiol. 2020 Feb;28(2):150-162. doi: 10.1016/j.tim.2019.09.002. Epub 2019 Oct 14. Trends Microbiol. 2020. PMID: 31624007 Free PMC article. Review.
-
Mutagenesis of the Varicella-Zoster Virus Genome Demonstrates That VLT and VLT-ORF63 Proteins Are Dispensable for Lytic Infection.Viruses. 2021 Nov 16;13(11):2289. doi: 10.3390/v13112289. Viruses. 2021. PMID: 34835095 Free PMC article.
-
The Presence of Antibody-Dependent Cell Cytotoxicity-Mediating Antibodies in Kaposi Sarcoma-Associated Herpesvirus-Seropositive Individuals Does Not Correlate with Disease Pathogenesis or Progression.J Immunol. 2020 Nov 15;205(10):2742-2749. doi: 10.4049/jimmunol.2000489. Epub 2020 Sep 30. J Immunol. 2020. PMID: 32998986 Free PMC article.
-
CRISPR/Cas9 ablating viral microRNA promotes lytic reactivation of Kaposi's sarcoma-associated herpesvirus.Biochem Biophys Res Commun. 2020 Dec 17;533(4):1400-1405. doi: 10.1016/j.bbrc.2020.10.030. Epub 2020 Oct 19. Biochem Biophys Res Commun. 2020. PMID: 33092788 Free PMC article.
References
-
- Bayley AC. 1984. Aggressive Kaposi's sarcoma in Zambia, 1983. Lancet 1:1318–1320. - PubMed
-
- Bourboulia D, Aldam D, Lagos D, Allen E, Williams I, Cornforth D, Copas A, Boshoff C. 2004. Short- and long-term effects of highly active antiretroviral therapy on Kaposi sarcoma-associated herpesvirus immune responses and viraemia. AIDS 18:485–493. doi:10.1097/00002030-200402200-00015. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

