Beyond transcription factors: roles of mRNA decay in regulating gene expression in plants
- PMID: 30613385
- PMCID: PMC6305221
- DOI: 10.12688/f1000research.16203.1
Beyond transcription factors: roles of mRNA decay in regulating gene expression in plants
Abstract
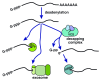
Gene expression is typically quantified as RNA abundance, which is influenced by both synthesis (transcription) and decay. Cytoplasmic decay typically initiates by deadenylation, after which decay can occur through any of three cytoplasmic decay pathways. Recent advances reveal several mechanisms by which RNA decay is regulated to control RNA abundance. mRNA can be post-transcriptionally modified, either indirectly through secondary structure or through direct modifications to the transcript itself, sometimes resulting in subsequent changes in mRNA decay rates. mRNA abundances can also be modified by tapping into pathways normally used for RNA quality control. Regulated mRNA decay can also come about through post-translational modification of decapping complex subunits. Likewise, mRNAs can undergo changes in subcellular localization (for example, the deposition of specific mRNAs into processing bodies, or P-bodies, where stabilization and destabilization occur in a transcript- and context-dependent manner). Additionally, specialized functions of mRNA decay pathways were implicated in a genome-wide mRNA decay analysis in Arabidopsis. Advances made using plants are emphasized in this review, but relevant studies from other model systems that highlight RNA decay mechanisms that may also be conserved in plants are discussed.
Keywords: DIS3L2; P-bodies; SOV; VCS; decapping; gene expression; mRNA decay.
Conflict of interest statement
No competing interests were disclosed.No competing interests were disclosed.No competing interests were disclosed.No competing interests were disclosed.
Figures

Similar articles
-
Arabidopsis mRNA decay landscape arises from specialized RNA decay substrates, decapping-mediated feedback, and redundancy.Proc Natl Acad Sci U S A. 2018 Feb 13;115(7):E1485-E1494. doi: 10.1073/pnas.1712312115. Epub 2018 Jan 31. Proc Natl Acad Sci U S A. 2018. PMID: 29386391 Free PMC article.
-
Kill the messenger: mRNA decay and plant development.Curr Opin Plant Biol. 2009 Feb;12(1):96-102. doi: 10.1016/j.pbi.2008.09.003. Epub 2008 Nov 5. Curr Opin Plant Biol. 2009. PMID: 18990607 Review.
-
mRNA decay in plants: both quantity and quality matter.Curr Opin Plant Biol. 2017 Feb;35:138-144. doi: 10.1016/j.pbi.2016.12.003. Epub 2016 Dec 20. Curr Opin Plant Biol. 2017. PMID: 28011423 Review.
-
Mechanisms of deadenylation-dependent decay.Wiley Interdiscip Rev RNA. 2011 Mar-Apr;2(2):167-83. doi: 10.1002/wrna.40. Epub 2010 Sep 15. Wiley Interdiscip Rev RNA. 2011. PMID: 21957004 Free PMC article. Review.
-
Targeted mRNA degradation by deadenylation-independent decapping.Mol Cell. 2004 Jul 2;15(1):5-15. doi: 10.1016/j.molcel.2004.06.028. Mol Cell. 2004. PMID: 15225544
Cited by
-
Genome-wide analysis of mRNA decay in Arabidopsis shoot and root reveals the importance of co-translational mRNA decay in the general mRNA turnover.Nucleic Acids Res. 2024 Jul 22;52(13):7910-7924. doi: 10.1093/nar/gkae363. Nucleic Acids Res. 2024. PMID: 38721772 Free PMC article.
-
RNA biology takes root in plant systems.Plant Direct. 2022 Sep 6;6(9):e445. doi: 10.1002/pld3.445. eCollection 2022 Sep. Plant Direct. 2022. PMID: 36091875 Free PMC article.
-
Inhibition of RNA degradation integrates the metabolic signals induced by osmotic stress into the Arabidopsis circadian system.J Exp Bot. 2023 Sep 29;74(18):5805-5819. doi: 10.1093/jxb/erad274. J Exp Bot. 2023. PMID: 37453132 Free PMC article.
-
What, where, and how: Regulation of translation and the translational landscape in plants.Plant Cell. 2024 May 1;36(5):1540-1564. doi: 10.1093/plcell/koad197. Plant Cell. 2024. PMID: 37437121 Free PMC article. Review.
-
RNA degradomes reveal substrates and importance for dark and nitrogen stress responses of Arabidopsis XRN4.Nucleic Acids Res. 2019 Sep 26;47(17):9216-9230. doi: 10.1093/nar/gkz712. Nucleic Acids Res. 2019. PMID: 31428786 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

