Synergy between RecBCD subunits is essential for efficient DNA unwinding
- PMID: 30601118
- PMCID: PMC6338465
- DOI: 10.7554/eLife.40836
Synergy between RecBCD subunits is essential for efficient DNA unwinding
Abstract
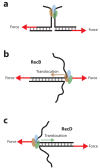
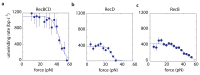
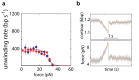
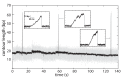
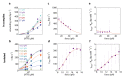
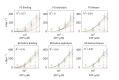
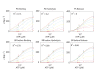
The subunits of the bacterial RecBCD act in coordination, rapidly and processively unwinding DNA at the site of a double strand break. RecBCD is able to displace DNA-binding proteins, suggesting that it generates high forces, but the specific role of each subunit in the force generation is unclear. Here, we present a novel optical tweezers assay that allows monitoring the activity of RecBCD's individual subunits, when they are part of an intact full complex. We show that RecBCD and its subunits are able to generate forces up to 25-40 pN without a significant effect on their velocity. Moreover, the isolated RecD translocates fast but is a weak helicase with limited processivity. Experiments at a broad range of [ATP] and forces suggest that RecD unwinds DNA as a Brownian ratchet, rectified by ATP binding, and that the presence of the other subunits shifts the ratchet equilibrium towards the post-translocation state.
Keywords: E. coli; RecBCD; biochemistry; chemical biology; helicase; molecular biophysics; optical tweezers; single molecule biophysics; structural biology.
© 2019, Zananiri et al.
Conflict of interest statement
RZ, OM, SR, VG, RK, AH, AK No competing interests declared
Figures



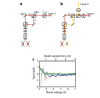
Similar articles
-
RecBCD enzyme is a bipolar DNA helicase.Nature. 2003 Jun 19;423(6942):893-7. doi: 10.1038/nature01673. Nature. 2003. PMID: 12815438
-
Bipolar DNA translocation contributes to highly processive DNA unwinding by RecBCD enzyme.J Biol Chem. 2005 Nov 4;280(44):37069-77. doi: 10.1074/jbc.M505520200. Epub 2005 Jul 22. J Biol Chem. 2005. PMID: 16041061
-
Sequence-dependent nanometer-scale conformational dynamics of individual RecBCD-DNA complexes.Nucleic Acids Res. 2016 Jul 8;44(12):5849-60. doi: 10.1093/nar/gkw445. Epub 2016 May 24. Nucleic Acids Res. 2016. PMID: 27220465 Free PMC article.
-
RecBCD is required to complete chromosomal replication: Implications for double-strand break frequencies and repair mechanisms.DNA Repair (Amst). 2015 Aug;32:86-95. doi: 10.1016/j.dnarep.2015.04.018. Epub 2015 May 2. DNA Repair (Amst). 2015. PMID: 26003632 Free PMC article. Review.
-
Single-strand gap repair involves both RecF and RecBCD pathways.Curr Genet. 2016 Aug;62(3):519-21. doi: 10.1007/s00294-016-0575-5. Epub 2016 Feb 13. Curr Genet. 2016. PMID: 26874520 Review.
Cited by
-
Three-Dimensional Tracking of Tethered Particles for Probing Nanometer-Scale Single-Molecule Dynamics Using a Plasmonic Microscope.ACS Sens. 2021 Nov 26;6(11):4234-4243. doi: 10.1021/acssensors.1c01927. Epub 2021 Nov 17. ACS Sens. 2021. PMID: 34786931 Free PMC article.
-
Single-molecule studies of helicases and translocases in prokaryotic genome-maintenance pathways.DNA Repair (Amst). 2021 Dec;108:103229. doi: 10.1016/j.dnarep.2021.103229. Epub 2021 Sep 20. DNA Repair (Amst). 2021. PMID: 34601381 Free PMC article. Review.
-
Communication between DNA and nucleotide binding sites facilitates stepping by the RecBCD helicase.Nucleic Acids Res. 2024 Apr 24;52(7):3911-3923. doi: 10.1093/nar/gkae108. Nucleic Acids Res. 2024. PMID: 38364872 Free PMC article.
-
A Tour de Force on the Double Helix: Exploiting DNA Mechanics To Study DNA-Based Molecular Machines.Biochemistry. 2019 Nov 26;58(47):4667-4676. doi: 10.1021/acs.biochem.9b00346. Epub 2019 Jun 28. Biochemistry. 2019. PMID: 31251042 Free PMC article. Review.
-
Insight into the biochemical mechanism of DNA helicases provided by bulk-phase and single-molecule assays.Methods. 2022 Aug;204:348-360. doi: 10.1016/j.ymeth.2021.12.002. Epub 2021 Dec 8. Methods. 2022. PMID: 34896247 Free PMC article. Review.
References
-
- Berg-Sørensen K, Flyvbjerg H. Power spectrum analysis for optical tweezers. Review of Scientific Instruments. 2004;75:594–612. doi: 10.1063/1.1645654. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

