Identification of the key genes and long non‑coding RNAs in ankylosing spondylitis using RNA sequencing
- PMID: 30592273
- PMCID: PMC6365023
- DOI: 10.3892/ijmm.2018.4038
Identification of the key genes and long non‑coding RNAs in ankylosing spondylitis using RNA sequencing
Erratum in
-
[Corrigendum] Identification of the key genes and long non‑coding RNAs in ankylosing spondylitis using RNA sequencing.Int J Mol Med. 2022 Jul;50(1):98. doi: 10.3892/ijmm.2022.5154. Epub 2022 Jun 1. Int J Mol Med. 2022. PMID: 35642663 Free PMC article.
Abstract
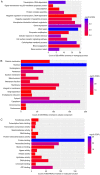
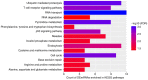
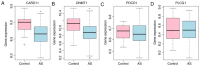
Ankylosing spondylitis (AS) is an insidious and debilitating form of arthritis that involves the axial skeleton, and its etiology and pathogenesis remain unclear. In the present study, three patients with AS and three normal controls from our hospital were enrolled. RNA sequencing and bioinformatics analysis were performed in order to identify the differentially expressed (DE) mRNAs (DEmRNAs) and DE long non‑coding RNAs (DElncRNAs) between the patients with AS and normal controls. Construction of an AS‑specific protein‑protein interaction network, a weighted DElncRNA‑DEmRNA co‑expression network and functional annotation of the DEmRNAs co‑expressed with DElncRNAs was performed. Nearby cis‑targeted DEmRNAs or DElncRNAs were identified by searching for DEmRNAs that were transcribed within 100‑kb up‑ or downstream of DElncRNAs. Based on the Gene Expression Omnibus datasets GSE25101 and GSE73754, the expression of selected DEmRNAs and DElncRNAs were verified using published RNA sequencing data from blood samples, and receiver operating characteristic analysis of selected DEmRNAs was performed. Compared with the normal controls, 1,072 DEmRNAs and 372 DElncRNAs in the patients with AS were identified. Caspase recruitment domain family member 11 and DNA methyltransferase 1 have great diagnostic value for AS. MSTRG.8559 and LINC00987 were also identified as two hub DElncRNAs. The T‑cell receptor signaling pathway was a significantly enriched pathway of the DEmRNAs co‑expressed with DElncRNAs in patients with AS. In conclusion, the present study identified the key DEmRNAs and DElncRNAs in AS, which provides novel information for understanding the pathogenesis of AS and developing potential biomarkers for AS.
Figures


Similar articles
-
Identification of aberrantly expressed long non-coding RNAs in postmenopausal osteoporosis.Int J Mol Med. 2018 Jun;41(6):3537-3550. doi: 10.3892/ijmm.2018.3575. Epub 2018 Mar 19. Int J Mol Med. 2018. PMID: 29568943 Free PMC article.
-
Screening key lncRNAs for human rectal adenocarcinoma based on lncRNA-mRNA functional synergistic network.Cancer Med. 2019 Jul;8(8):3875-3891. doi: 10.1002/cam4.2236. Epub 2019 May 22. Cancer Med. 2019. PMID: 31116002 Free PMC article.
-
Identification of aberrantly expressed long non-coding RNAs in stomach adenocarcinoma.Oncotarget. 2017 Jul 25;8(30):49201-49216. doi: 10.18632/oncotarget.17329. Oncotarget. 2017. PMID: 28484081 Free PMC article.
-
Identification and bioinformatics analysis of lncRNAs in serum of patients with ankylosing spondylitis.BMC Musculoskelet Disord. 2024 Apr 15;25(1):291. doi: 10.1186/s12891-024-07396-z. BMC Musculoskelet Disord. 2024. PMID: 38622662 Free PMC article. Review.
-
Reconstruction and Analysis of the Differentially Expressed IncRNA-miRNA-mRNA Network Based on Competitive Endogenous RNA in Hepatocellular Carcinoma.Crit Rev Eukaryot Gene Expr. 2019;29(6):539-549. doi: 10.1615/CritRevEukaryotGeneExpr.2019028740. Crit Rev Eukaryot Gene Expr. 2019. PMID: 32422009 Review.
Cited by
-
Emerging Roles of Long Non-Coding RNAs in Ankylosing Spondylitis.Front Immunol. 2022 Feb 10;13:790924. doi: 10.3389/fimmu.2022.790924. eCollection 2022. Front Immunol. 2022. PMID: 35222376 Free PMC article. Review.
-
H19 Increases IL-17A/IL-23 Releases via Regulating VDR by Interacting with miR675-5p/miR22-5p in Ankylosing Spondylitis.Mol Ther Nucleic Acids. 2020 Mar 6;19:393-404. doi: 10.1016/j.omtn.2019.11.025. Epub 2019 Nov 29. Mol Ther Nucleic Acids. 2020. PMID: 31887550 Free PMC article.
-
Comparative transcriptome analysis of transcripts of uncertain coding potential in septic myocardial depression.BMC Cardiovasc Disord. 2021 Apr 8;21(1):166. doi: 10.1186/s12872-021-01973-z. BMC Cardiovasc Disord. 2021. PMID: 33832434 Free PMC article.
-
MicroRNA let-7i-3p affects osteoblast differentiation in ankylosing spondylitis via targeting PDK1.Cell Cycle. 2021 Jun;20(12):1209-1219. doi: 10.1080/15384101.2021.1930680. Epub 2021 May 28. Cell Cycle. 2021. PMID: 34048311 Free PMC article.
-
LncRNA MEG3 inhibits the inflammatory response of ankylosing spondylitis by targeting miR-146a.Mol Cell Biochem. 2020 Mar;466(1-2):17-24. doi: 10.1007/s11010-019-03681-x. Epub 2020 Jan 1. Mol Cell Biochem. 2020. PMID: 31894531
References
-
- Evans DM, Spencer CC, Pointon JJ, Su Z, Harvey D, Kochan G, Oppermann U, Dilthey A, Pirinen M, Stone MA, et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat Genet. 2011;43:761–767. doi: 10.1038/ng.873. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

