LINE-1 and EPAS1 DNA methylation associations with high-altitude exposure
- PMID: 30574831
- PMCID: PMC6380404
- DOI: 10.1080/15592294.2018.1561117
LINE-1 and EPAS1 DNA methylation associations with high-altitude exposure
Abstract
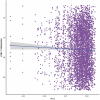
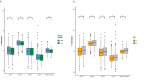
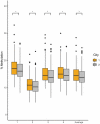
Recent discoveries indicate a genetic basis for high-altitude adaptation among human groups who have resided at high altitude for millennia, including Andeans, Tibetans, and Ethiopians. Yet, genetics alone does not explain the extent of variation in altitude-adaptive phenotypes. Current and past environments may also play a role, and one way to determine the effect of the environment is through the epigenome. To characterize if Andean adaptive responses to high altitude have an epigenetic component, we analyzed DNA methylation of the promoter region of EPAS1 and LINE-1 repetitive element among 572 Quechua individuals from high- (4,388 m) and low-altitude (0 m) in Peru. Participants recruited at high altitude had lower EPAS1 DNA methylation and higher LINE-1 methylation. Altitude of birth was associated with higher LINE-1 methylation, not with EPAS1 methylation. The number of years lived at high altitude was negatively associated with EPAS1 methylation and positively associated with LINE-1 methylation. We found four one-carbon metabolism SNPs (MTHFD1 rs2236225, TYMS rs502396, FOLH1 rs202676, GLDC rs10975681) that cumulatively explained 11.29% of the variation in average LINE-1 methylation. And identified an association between LINE-1 methylation and genome-wide SNP principal component 1 that distinguishes European from Indigenous American ancestry suggesting that European admixture decreases LINE-1 methylation. Our results indicate that both current and lifetime exposure to high-altitude hypoxia have an effect on EPAS1 and LINE-1 methylation among Andean Quechua, suggesting that epigenetic modifications may play a role in high-altitude adaptation.
Keywords: Andes; DNA methylation; Epigenetics; adaptation; hypoxia.
Figures
Similar articles
-
Genome-Wide Epigenetic Signatures of Adaptive Developmental Plasticity in the Andes.Genome Biol Evol. 2021 Feb 3;13(2):evaa239. doi: 10.1093/gbe/evaa239. Genome Biol Evol. 2021. PMID: 33185669 Free PMC article.
-
Down-Regulation of EPAS1 Transcription and Genetic Adaptation of Tibetans to High-Altitude Hypoxia.Mol Biol Evol. 2017 Apr 1;34(4):818-830. doi: 10.1093/molbev/msw280. Mol Biol Evol. 2017. PMID: 28096303 Free PMC article.
-
Physiological and genomic evidence that selection on the transcription factor Epas1 has altered cardiovascular function in high-altitude deer mice.PLoS Genet. 2019 Nov 7;15(11):e1008420. doi: 10.1371/journal.pgen.1008420. eCollection 2019 Nov. PLoS Genet. 2019. PMID: 31697676 Free PMC article.
-
Metabolic aspects of high-altitude adaptation in Tibetans.Exp Physiol. 2015 Nov;100(11):1247-55. doi: 10.1113/EP085292. Epub 2015 Jul 14. Exp Physiol. 2015. PMID: 26053282 Free PMC article. Review.
-
Human high-altitude adaptation: forward genetics meets the HIF pathway.Genes Dev. 2014 Oct 15;28(20):2189-204. doi: 10.1101/gad.250167.114. Genes Dev. 2014. PMID: 25319824 Free PMC article. Review.
Cited by
-
Hypoxia-induced signaling in the cardiovascular system: pathogenesis and therapeutic targets.Signal Transduct Target Ther. 2023 Nov 20;8(1):431. doi: 10.1038/s41392-023-01652-9. Signal Transduct Target Ther. 2023. PMID: 37981648 Free PMC article. Review.
-
Genome-Wide Epigenetic Signatures of Adaptive Developmental Plasticity in the Andes.Genome Biol Evol. 2021 Feb 3;13(2):evaa239. doi: 10.1093/gbe/evaa239. Genome Biol Evol. 2021. PMID: 33185669 Free PMC article.
-
Time Domains of Hypoxia Responses and -Omics Insights.Front Physiol. 2022 Aug 8;13:885295. doi: 10.3389/fphys.2022.885295. eCollection 2022. Front Physiol. 2022. PMID: 36035495 Free PMC article. Review.
-
Notch Signaling and Cross-Talk in Hypoxia: A Candidate Pathway for High-Altitude Adaptation.Life (Basel). 2022 Mar 16;12(3):437. doi: 10.3390/life12030437. Life (Basel). 2022. PMID: 35330188 Free PMC article. Review.
-
An Aptitude for Altitude: Are Epigenomic Processes Involved?Front Physiol. 2019 Nov 22;10:1397. doi: 10.3389/fphys.2019.01397. eCollection 2019. Front Physiol. 2019. PMID: 31824328 Free PMC article. Review.
References
-
- Moore LG. Human genetic adaptation to high altitude. High Alt Med Biol. 2001. Summer;2(2):257–279. PubMed PMID: 11443005. - PubMed
-
- Bigham AW, Julian CG, Wilson MJ, et al. Maternal PRKAA1 and EDNRA genotypes are associated with birth weight, and PRKAA1 with uterine artery diameter and metabolic homeostasis at high altitude. Physiol Genomics. 2014. September 15;46(18):687–697. PubMed PMID: 25225183; PubMed Central PMCID: PMC4166715. - PMC - PubMed
-
- Simonson TS, Yang Y, Huff CD, et al. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010. July 2;329(5987):72–75. PubMed PMID: 20466884. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
