Exploitation of Synthetic mRNA To Drive Immune Effector Cell Recruitment and Functional Reprogramming In Vivo
- PMID: 30541883
- PMCID: PMC6325005
- DOI: 10.4049/jimmunol.1800924
Exploitation of Synthetic mRNA To Drive Immune Effector Cell Recruitment and Functional Reprogramming In Vivo
Abstract
Therapeutic strategies based on in vitro-transcribed mRNA (IVT) are attractive because they avoid the permanent signature of genomic integration that is associated with DNA-based therapy and result in the transient production of proteins of interest. To date, IVT has mainly been used in vaccination protocols to generate immune responses to foreign Ags. In this "proof-of-principle" study, we explore a strategy of combinatorial IVT to recruit and reprogram immune effector cells to acquire divergent biological functions in mice in vivo. First, we demonstrate that synthetic mRNA encoding CCL3 is able to recruit murine monocytes in a nonprogrammed state, exhibiting neither bactericidal nor tissue-repairing properties. However, upon addition of either Ifn-γ mRNA or Il-4 mRNA, we successfully polarized these cells to adopt either M1 or M2 macrophage activation phenotypes. This cellular reprogramming was demonstrated through increased expression of known surface markers and through the differential modulation of NADPH oxidase activity, or the superoxide burst. Our study demonstrates how IVT strategies can be combined to recruit and reprogram immune effector cells that have the capacity to fulfill complex biological tasks in vivo.
Copyright © 2019 by The American Association of Immunologists, Inc.
Figures
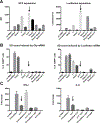
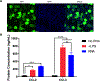

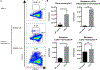
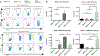


Similar articles
-
Genetically Modified Live Attenuated Leishmania donovani Parasites Induce Innate Immunity through Classical Activation of Macrophages That Direct the Th1 Response in Mice.Infect Immun. 2015 Oct;83(10):3800-15. doi: 10.1128/IAI.00184-15. Epub 2015 Jul 13. Infect Immun. 2015. PMID: 26169275 Free PMC article.
-
Innate cellular sources of interleukin-17A regulate macrophage accumulation in cigarette- smoke-induced lung inflammation in mice.Clin Sci (Lond). 2015 Nov;129(9):785-96. doi: 10.1042/CS20140703. Epub 2015 Jul 1. Clin Sci (Lond). 2015. PMID: 26201093 Free PMC article.
-
Temporal phenotypic features distinguish polarized macrophages in vitro.Autoimmunity. 2015 May;48(3):161-76. doi: 10.3109/08916934.2015.1027816. Epub 2015 Mar 31. Autoimmunity. 2015. PMID: 25826285 Free PMC article.
-
Concise Review: Application of In Vitro Transcribed Messenger RNA for Cellular Engineering and Reprogramming: Progress and Challenges.Stem Cells. 2017 Jan;35(1):68-79. doi: 10.1002/stem.2402. Epub 2016 Jun 20. Stem Cells. 2017. PMID: 27250673 Review.
-
Advancing in vivo reprogramming with synthetic biology.Curr Opin Biotechnol. 2024 Jun;87:103109. doi: 10.1016/j.copbio.2024.103109. Epub 2024 Mar 24. Curr Opin Biotechnol. 2024. PMID: 38520824 Free PMC article. Review.
Cited by
-
Engineering monoclonal antibody-based contraception and multipurpose prevention technologies†.Biol Reprod. 2020 Aug 4;103(2):275-285. doi: 10.1093/biolre/ioaa096. Biol Reprod. 2020. PMID: 32607584 Free PMC article. Review.
-
Evaluation of M2-like macrophage enrichment after diffuse traumatic brain injury through transient interleukin-4 expression from engineered mesenchymal stromal cells.J Neuroinflammation. 2020 Jun 20;17(1):197. doi: 10.1186/s12974-020-01860-y. J Neuroinflammation. 2020. PMID: 32563258 Free PMC article.
-
Chemical modification of uridine modulates mRNA-mediated proinflammatory and antiviral response in primary human macrophages.Mol Ther Nucleic Acids. 2022 Jan 10;27:854-869. doi: 10.1016/j.omtn.2022.01.004. eCollection 2022 Mar 8. Mol Ther Nucleic Acids. 2022. PMID: 35141046 Free PMC article.
-
Leveraging immune memory against measles virus as an antitumor strategy in a preclinical model of aggressive squamous cell carcinoma.J Immunother Cancer. 2021 Oct;9(10):e002170. doi: 10.1136/jitc-2020-002170. J Immunother Cancer. 2021. PMID: 34675067 Free PMC article.
-
Cell Reprogramming and Differentiation Utilizing Messenger RNA for Regenerative Medicine.J Dev Biol. 2023 Dec 20;12(1):1. doi: 10.3390/jdb12010001. J Dev Biol. 2023. PMID: 38535481 Free PMC article. Review.
References
-
- Kuhn AN, Beibetaert T, Simon P, Vallazza B, Buck J, Davies BP, Tureci O, and Sahin U. 2012. mRNA as a versatile tool for exogenous protein expression. Curr Gene Ther 12: 347–361. - PubMed
-
- Sahin U, Kariko K, and Tureci O. 2014. mRNA-based therapeutics--developing a new class of drugs. Nat Rev Drug Discov 13: 759–780. - PubMed
-
- Kormann MS, Hasenpusch G, Aneja MK, Nica G, Flemmer AW, Herber-Jonat S, Huppmann M, Mays LE, Illenyi M, Schams A, Griese M, Bittmann I, Handgretinger R, Hartl D, Rosenecker J, and Rudolph C. 2011. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat Biotechnol 29: 154–157. - PubMed
-
- Andries O, Mc Cafferty S, De Smedt SC, Weiss R, Sanders NN, and Kitada T. 2015. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J Control Release 217: 337–344. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

