Autophagy Promotes Replication of Influenza A Virus In Vitro
- PMID: 30541828
- PMCID: PMC6363991
- DOI: 10.1128/JVI.01984-18
Autophagy Promotes Replication of Influenza A Virus In Vitro
Abstract
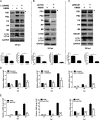
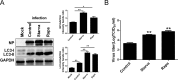
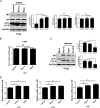
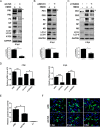
Influenza A virus (IAV) infection could induce autophagosome accumulation. However, the impact of the autophagy machinery on IAV infection remains controversial. Here, we showed that induction of cellular autophagy by starvation or rapamycin treatment increases progeny virus production, while disruption of autophagy using a small interfering RNA (siRNA) and pharmacological inhibitor reduces progeny virus production. Further studies revealed that alteration of autophagy significantly affects the early stages of the virus life cycle or viral RNA synthesis. Importantly, we demonstrated that overexpression of both the IAV M2 and NP proteins alone leads to the lipidation of LC3 to LC3-II and a redistribution of LC3 from the cytosol to punctate vesicles indicative of authentic autophagosomes. Intriguingly, both M2 and NP colocalize and interact with LC3 puncta during M2 or NP transfection alone and IAV infection, leading to an increase in viral ribonucleoprotein (vRNP) export and infectious viral particle formation, which indicates that the IAV-host autophagy interaction plays a critical role in regulating IAV replication. We showed that NP and M2 induce the AKT-mTOR-dependent autophagy pathway and an increase in HSP90AA1 expression. Finally, our studies provided evidence that IAV replication needs an autophagy pathway to enhance viral RNA synthesis via the interaction of PB2 and HSP90AA1 by modulating HSP90AA1 expression and the AKT-mTOR signaling pathway in host cells. Collectively, our studies uncover a new mechanism that NP- and M2-mediated autophagy functions in different stages of virus replication in the pathogenicity of influenza A virus.IMPORTANCE Autophagy impacts the replication cycle of many viruses. However, the role of the autophagy machinery in IAV replication remains unclear. Therefore, we explored the detailed mechanisms utilized by IAV to promote its replication. We demonstrated that IAV NP- and M2-mediated autophagy promotes IAV replication by regulating the AKT-mTOR signaling pathway and HSP90AA1 expression. The interaction of PB2 and HSP90AA1 results in the increase of viral RNA synthesis first; subsequently the binding of NP to LC3 favors vRNP export, and later the interaction of M2 and LC3 leads to an increase in the production of infectious viral particles, thus accelerating viral progeny production. These findings improve our understanding of IAV pathogenicity in host cells.
Keywords: HSP90AA1; IAV; LC3; M2; NP; autophagy; replication.
Copyright © 2019 American Society for Microbiology.
Figures



Similar articles
-
Y-Box-Binding Protein 3 (YBX3) Restricts Influenza A Virus by Interacting with Viral Ribonucleoprotein Complex and Imparing its Function.J Gen Virol. 2020 Apr;101(4):385-398. doi: 10.1099/jgv.0.001390. J Gen Virol. 2020. PMID: 32553055
-
Eukaryotic Translation Elongation Factor 1 Delta Inhibits the Nuclear Import of the Nucleoprotein and PA-PB1 Heterodimer of Influenza A Virus.J Virol. 2020 Dec 22;95(2):e01391-20. doi: 10.1128/JVI.01391-20. Print 2020 Dec 22. J Virol. 2020. PMID: 33087462 Free PMC article.
-
The Nucleolar Protein LYAR Facilitates Ribonucleoprotein Assembly of Influenza A Virus.J Virol. 2018 Nov 12;92(23):e01042-18. doi: 10.1128/JVI.01042-18. Print 2018 Dec 1. J Virol. 2018. PMID: 30209172 Free PMC article.
-
The battle for autophagy between host and influenza A virus.Virulence. 2022 Dec;13(1):46-59. doi: 10.1080/21505594.2021.2014680. Virulence. 2022. PMID: 34967267 Free PMC article. Review.
-
Biogenesis, assembly, and export of viral messenger ribonucleoproteins in the influenza A virus infected cell.RNA Biol. 2013 Aug;10(8):1274-82. doi: 10.4161/rna.25356. Epub 2013 Jun 17. RNA Biol. 2013. PMID: 23807439 Free PMC article. Review.
Cited by
-
Functions of Viroporins in the Viral Life Cycle and Their Regulation of Host Cell Responses.Front Immunol. 2022 Jun 2;13:890549. doi: 10.3389/fimmu.2022.890549. eCollection 2022. Front Immunol. 2022. PMID: 35720341 Free PMC article. Review.
-
Unravelling the Immunomodulatory Effects of Viral Ion Channels, towards the Treatment of Disease.Viruses. 2021 Oct 27;13(11):2165. doi: 10.3390/v13112165. Viruses. 2021. PMID: 34834972 Free PMC article. Review.
-
Simultaneous co-infection with swine influenza A and porcine reproductive and respiratory syndrome viruses potentiates adaptive immune responses.Front Immunol. 2023 May 23;14:1192604. doi: 10.3389/fimmu.2023.1192604. eCollection 2023. Front Immunol. 2023. PMID: 37287962 Free PMC article.
-
Porcine reproductive and respiratory syndrome virus nonstructural protein 2 promotes the autophagic degradation of adaptor protein SH3KBP1 to antagonize host innate immune responses by enhancing K63-linked polyubiquitination of RIG-I.PLoS Pathog. 2024 Oct 28;20(10):e1012670. doi: 10.1371/journal.ppat.1012670. eCollection 2024 Oct. PLoS Pathog. 2024. PMID: 39466846 Free PMC article.
-
Cellular Proteostasis During Influenza A Virus Infection-Friend or Foe?Cells. 2019 Mar 9;8(3):228. doi: 10.3390/cells8030228. Cells. 2019. PMID: 30857287 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

