RNF126 Quenches RNF168 Function in the DNA Damage Response
- PMID: 30529286
- PMCID: PMC6411902
- DOI: 10.1016/j.gpb.2018.07.004
RNF126 Quenches RNF168 Function in the DNA Damage Response
Abstract
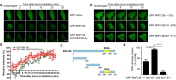
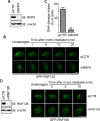
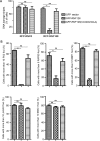
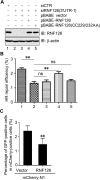
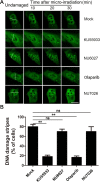
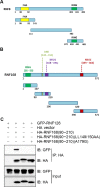
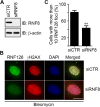
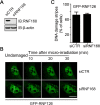
DNA damage response (DDR) is essential for maintaining genome stability and protecting cells from tumorigenesis. Ubiquitin and ubiquitin-like modifications play an important role in DDR, from signaling DNA damage to mediating DNA repair. In this report, we found that the E3 ligase ring finger protein 126 (RNF126) was recruited to UV laser micro-irradiation-induced stripes in a RNF8-dependent manner. RNF126 directly interacted with and ubiquitinated another E3 ligase, RNF168. Overexpression of wild type RNF126, but not catalytically-inactive mutant RNF126 (CC229/232AA), diminished ubiquitination of H2A histone family member X (H2AX), and subsequent bleomycin-induced focus formation of total ubiquitin FK2, TP53-binding protein 1 (53BP1), and receptor-associated protein 80 (RAP80). Interestingly, both RNF126 overexpression and RNF126 downregulation compromised homologous recombination (HR)-mediated repair of DNA double-strand breaks (DSBs). Taken together, our findings demonstrate that RNF126 negatively regulates RNF168 function in DDR and its appropriate cellular expression levels are essential for HR-mediated DSB repair.
Keywords: DNA damage response; DNA repair; RNF126; RNF168; RNF8; Ubiquitination.
Copyright © 2018 The Authors. Production and hosting by Elsevier B.V. All rights reserved.
Figures


Similar articles
-
Ring finger protein 126 (RNF126) suppresses ionizing radiation-induced p53-binding protein 1 (53BP1) focus formation.J Biol Chem. 2018 Jan 12;293(2):588-598. doi: 10.1074/jbc.M116.765602. Epub 2017 Nov 22. J Biol Chem. 2018. PMID: 29167269 Free PMC article.
-
Human RNF169 is a negative regulator of the ubiquitin-dependent response to DNA double-strand breaks.J Cell Biol. 2012 Apr 16;197(2):189-99. doi: 10.1083/jcb.201109100. Epub 2012 Apr 9. J Cell Biol. 2012. PMID: 22492721 Free PMC article.
-
USP14 regulates DNA damage repair by targeting RNF168-dependent ubiquitination.Autophagy. 2018;14(11):1976-1990. doi: 10.1080/15548627.2018.1496877. Epub 2018 Aug 10. Autophagy. 2018. PMID: 29995557 Free PMC article.
-
Opposing roles of RNF8/RNF168 and deubiquitinating enzymes in ubiquitination-dependent DNA double-strand break response signaling and DNA-repair pathway choice.J Radiat Res. 2016 Aug;57 Suppl 1(Suppl 1):i33-i40. doi: 10.1093/jrr/rrw027. Epub 2016 Mar 16. J Radiat Res. 2016. PMID: 26983989 Free PMC article. Review.
-
RAP80 and RNF8, key players in the recruitment of repair proteins to DNA damage sites.Cancer Lett. 2008 Nov 28;271(2):179-90. doi: 10.1016/j.canlet.2008.04.046. Epub 2008 Jun 11. Cancer Lett. 2008. PMID: 18550271 Free PMC article. Review.
Cited by
-
Role of RING-Type E3 Ubiquitin Ligases in Inflammatory Signalling and Inflammatory Bowel Disease.Mediators Inflamm. 2020 Aug 10;2020:5310180. doi: 10.1155/2020/5310180. eCollection 2020. Mediators Inflamm. 2020. PMID: 32848509 Free PMC article. Review.
-
RNF126-Mediated MRE11 Ubiquitination Activates the DNA Damage Response and Confers Resistance of Triple-Negative Breast Cancer to Radiotherapy.Adv Sci (Weinh). 2023 Feb;10(5):e2203884. doi: 10.1002/advs.202203884. Epub 2022 Dec 23. Adv Sci (Weinh). 2023. PMID: 36563124 Free PMC article.
-
Molecular Mechanisms of Arsenic-Induced Disruption of DNA Repair.Chem Res Toxicol. 2020 Mar 16;33(3):709-726. doi: 10.1021/acs.chemrestox.9b00464. Epub 2020 Feb 7. Chem Res Toxicol. 2020. PMID: 31986875 Free PMC article. Review.
-
Regulation of chromatin structure and function: insights into the histone chaperone FACT.Cell Cycle. 2021 Mar-Mar;20(5-6):465-479. doi: 10.1080/15384101.2021.1881726. Epub 2021 Feb 16. Cell Cycle. 2021. PMID: 33590780 Free PMC article. Review.
-
Ubiquitin E3 ligases in cancer: somatic mutation and amplification.BMB Rep. 2023 May;56(5):265-274. doi: 10.5483/BMBRep.2023-0037. BMB Rep. 2023. PMID: 37081755 Free PMC article. Review.
References
-
- Carrassa L., Damia G. DNA damage response inhibitors: mechanisms and potential applications in cancer therapy. Cancer Treat Rev. 2017;60:139–151. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

