Enzymatic Phosphorylation of Ser in a Type I Collagen Peptide
- PMID: 30527445
- PMCID: PMC6302033
- DOI: 10.1016/j.bpj.2018.11.012
Enzymatic Phosphorylation of Ser in a Type I Collagen Peptide
Abstract
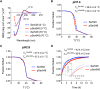
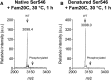
Phosphoproteomics studies have reported phosphorylation at multiple sites within collagen, raising the possibility that these post-translational modifications regulate the physical or biological properties of collagen. In this study, molecular dynamics simulations and experimental studies were carried out on model peptides to establish foundational principles of phosphorylation of Ser residues in collagen. A (Gly-Xaa-Yaa)11 peptide was designed to include a Ser-containing sequence from type I collagen that was reported to be phosphorylated. The physiological kinase involved in collagen phosphorylation is not known. In vitro studies showed that a model kinase ERK1 (extracellular signal-regulated protein kinase 1) would phosphorylate Ser within the consensus sequence if the collagen-like peptide is in the denatured state but not in the triple-helical state. The peptide was not a substrate for FAM20C, a kinase present in the secretory pathway, which has been shown to phosphorylate many extracellular matrix proteins. The unfolded single chain (Gly-Xaa-Yaa)11 peptide containing phosphoSer was able to refold to form a stable triple helix but at a reduced folding rate and with a small decrease in thermal stability relative to the nonphosphorylated peptide at neutral pH. These biophysical studies on model peptides provide a basis for investigations into the physiological consequences of collagen phosphorylation and the application of phosphorylation to regulate the properties of collagen biomaterials.
Copyright © 2018 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
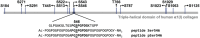




Similar articles
-
Amino acid sequence environment modulates the disruption by osteogenesis imperfecta glycine substitutions in collagen-like peptides.Biochemistry. 1997 Jun 10;36(23):6930-5. doi: 10.1021/bi970051h. Biochemistry. 1997. PMID: 9188687
-
Consequences of Glycine Mutations in the Fibronectin-binding Sequence of Collagen.J Biol Chem. 2016 Dec 30;291(53):27073-27086. doi: 10.1074/jbc.M116.753566. Epub 2016 Oct 31. J Biol Chem. 2016. PMID: 27799304 Free PMC article.
-
Stabilization of triple-helical structures of collagen peptides containing a Hyp-Thr-Gly, Hyp-Val-Gly, or Hyp-Ser-Gly sequence.Biopolymers. 2011 Sep;95(9):628-40. doi: 10.1002/bip.21625. Epub 2011 Mar 25. Biopolymers. 2011. PMID: 21442606
-
Triple-helical peptides: an approach to collagen conformation, stability, and self-association.Biopolymers. 2008 May;89(5):345-53. doi: 10.1002/bip.20958. Biopolymers. 2008. PMID: 18275087 Review.
-
The crucial role of trimerization domains in collagen folding.Int J Biochem Cell Biol. 2012 Jan;44(1):21-32. doi: 10.1016/j.biocel.2011.09.009. Epub 2011 Oct 5. Int J Biochem Cell Biol. 2012. PMID: 22001560 Review.
Cited by
-
The ABCs of the atypical Fam20 secretory pathway kinases.J Biol Chem. 2021 Jan-Jun;296:100267. doi: 10.1016/j.jbc.2021.100267. Epub 2021 Jan 8. J Biol Chem. 2021. PMID: 33759783 Free PMC article. Review.
-
Fam20C in Human Diseases: Emerging Biological Functions and Therapeutic Implications.Front Mol Biosci. 2021 Dec 20;8:790172. doi: 10.3389/fmolb.2021.790172. eCollection 2021. Front Mol Biosci. 2021. PMID: 34988120 Free PMC article. Review.
-
Analysis of pancreatic extracellular matrix protein post-translational modifications via electrostatic repulsion-hydrophilic interaction chromatography coupled with mass spectrometry.Mol Omics. 2021 Oct 11;17(5):652-664. doi: 10.1039/d1mo00104c. Mol Omics. 2021. PMID: 34318855 Free PMC article.
-
Clinical and functional characterization of COL2A1 p.Gly444Ser variant: From a fetal phenotype to a previously undisclosed postnatal phenotype.Bone Rep. 2023 Nov 27;19:101728. doi: 10.1016/j.bonr.2023.101728. eCollection 2023 Dec. Bone Rep. 2023. PMID: 38076483 Free PMC article.
-
Structural basis for the hyperthermostability of an archaeal enzyme induced by succinimide formation.Biophys J. 2021 Sep 7;120(17):3732-3746. doi: 10.1016/j.bpj.2021.07.014. Epub 2021 Jul 22. Biophys J. 2021. PMID: 34302792 Free PMC article.
References
-
- Bella J. Collagen structure: new tricks from a very old dog. Biochem. J. 2016;473:1001–1025. - PubMed
-
- Brodsky B., Persikov A.V. Molecular structure of the collagen triple helix. Adv. Protein Chem. 2005;70:301–339. - PubMed
-
- Kramer R.Z., Venugopal M.G., Berman H.M. Staggered molecular packing in crystals of a collagen-like peptide with a single charged pair. J. Mol. Biol. 2000;301:1191–1205. - PubMed
-
- Burjanadze T.V. Hydroxyproline content and location in relation to collagen thermal stability. Biopolymers. 1979;18:931–938. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

