A DNA methylation reader complex that enhances gene transcription
- PMID: 30523112
- PMCID: PMC6353633
- DOI: 10.1126/science.aar7854
A DNA methylation reader complex that enhances gene transcription
Abstract
DNA methylation generally functions as a repressive transcriptional signal, but it is also known to activate gene expression. In either case, the downstream factors remain largely unknown. By using comparative interactomics, we isolated proteins in Arabidopsis thaliana that associate with methylated DNA. Two SU(VAR)3-9 homologs, the transcriptional antisilencing factor SUVH1, and SUVH3, were among the methyl reader candidates. SUVH1 and SUVH3 bound methylated DNA in vitro, were associated with euchromatic methylation in vivo, and formed a complex with two DNAJ domain-containing homologs, DNAJ1 and DNAJ2. Ectopic recruitment of DNAJ1 enhanced gene transcription in plants, yeast, and mammals. Thus, the SUVH proteins bind to methylated DNA and recruit the DNAJ proteins to enhance proximal gene expression, thereby counteracting the repressive effects of transposon insertion near genes.
Copyright © 2018 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures


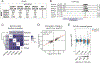

Comment in
-
Freedom of expression in methylated regions.Nat Rev Mol Cell Biol. 2019 Feb;20(2):66-67. doi: 10.1038/s41580-018-0097-8. Nat Rev Mol Cell Biol. 2019. PMID: 30575802 No abstract available.
Similar articles
-
Intron-regulated expression of SUVH3, an Arabidopsis Su(var)3-9 homologue.J Exp Bot. 2006;57(12):3301-11. doi: 10.1093/jxb/erl093. Epub 2006 Aug 23. J Exp Bot. 2006. PMID: 16928780
-
A methylated-DNA-binding complex required for plant development mediates transcriptional activation of promoter methylated genes.J Integr Plant Biol. 2019 Feb;61(2):120-139. doi: 10.1111/jipb.12767. J Integr Plant Biol. 2019. PMID: 30589221
-
SUVH1, a Su(var)3-9 family member, promotes the expression of genes targeted by DNA methylation.Nucleic Acids Res. 2016 Jan 29;44(2):608-20. doi: 10.1093/nar/gkv958. Epub 2015 Sep 22. Nucleic Acids Res. 2016. PMID: 26400170 Free PMC article.
-
Heterochromatin proteins and the control of heterochromatic gene silencing in Arabidopsis.J Plant Physiol. 2006 Feb;163(3):358-68. doi: 10.1016/j.jplph.2005.10.015. Epub 2005 Dec 27. J Plant Physiol. 2006. PMID: 16384625 Review.
-
Novel DNAJ-related proteins in Arabidopsis thaliana.New Phytol. 2018 Jan;217(2):480-490. doi: 10.1111/nph.14827. New Phytol. 2018. PMID: 29271039 Review.
Cited by
-
Perspectives for epigenetic editing in crops.Transgenic Res. 2021 Aug;30(4):381-400. doi: 10.1007/s11248-021-00252-z. Epub 2021 Apr 23. Transgenic Res. 2021. PMID: 33891288 Review.
-
Population-level analysis reveals the widespread occurrence and phenotypic consequence of DNA methylation variation not tagged by genetic variation in maize.Genome Biol. 2019 Nov 19;20(1):243. doi: 10.1186/s13059-019-1859-0. Genome Biol. 2019. PMID: 31744513 Free PMC article.
-
Genome-wide analysis of short interspersed nuclear elements provides insight into gene and genome evolution in citrus.DNA Res. 2020 Feb 1;27(1):dsaa004. doi: 10.1093/dnares/dsaa004. DNA Res. 2020. PMID: 32271875 Free PMC article.
-
Chromatin regulation in plant hormone and plant stress responses.Curr Opin Plant Biol. 2020 Oct;57:164-170. doi: 10.1016/j.pbi.2020.08.007. Epub 2020 Nov 2. Curr Opin Plant Biol. 2020. PMID: 33142261 Free PMC article. Review.
-
The expanding world of plant J-domain proteins.CRC Crit Rev Plant Sci. 2019;38(5-6):382-400. doi: 10.1080/07352689.2019.1693716. Epub 2019 Dec 10. CRC Crit Rev Plant Sci. 2019. PMID: 33223602 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

