Genome-wide identification, molecular evolution, and expression analysis of auxin response factor (ARF) gene family in Brachypodium distachyon L
- PMID: 30522432
- PMCID: PMC6282295
- DOI: 10.1186/s12870-018-1559-z
Genome-wide identification, molecular evolution, and expression analysis of auxin response factor (ARF) gene family in Brachypodium distachyon L
Abstract
Background: The auxin response factor (ARF) gene family is involved in plant development and hormone regulation. Although the ARF gene family has been studied in some plant species, its structural features, molecular evolution, and expression profiling in Brachypodium distachyon L. are still not clear.
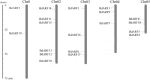
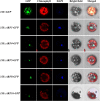
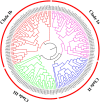
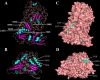
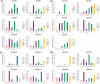
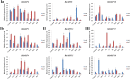
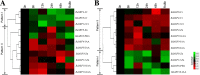
Results: Genome-wide analysis identified 19 ARF genes in B. distachyon. A phylogenetic tree constructed with 182 ARF genes from seven plant species revealed three different clades, and the ARF genes from within a clade exhibited structural conservation, although certain divergences occurred in different clades. The branch-site model identified some sites where positive selection may have occurred, and functional divergence analysis found more Type II divergence sites than Type I. In particular, both positive selection and functional divergence may have occurred in 241H, 243G, 244 L, 310 T, 340G and 355 T. Subcellular localization prediction and experimental verification indicated that BdARF proteins were present in the nucleus. Transcript expression analysis revealed that BdARFs were mainly expressed in the leaf and root tips, stems, and developing seeds. Some BdARF genes exhibited significantly upregulated expression under various abiotic stressors. Particularly, BdARF4 and BdARF8 were significantly upregulated in response to abiotic stress factors such as salicylic acid and heavy metals.
Conclusion: The ARF gene family in B. distachyon was highly conserved. Several important amino acid sites were identified where positive selection and functional divergence occurred, and they may play important roles in functional differentiation. BdARF genes had clear tissue and organ expression preference and were involved in abiotic stress response, suggesting their roles in plant growth and stress resistance.
Keywords: ARF genes; Abiotic stress; Brachypodium distachyon; Phylogenetic relationships; qRT-PCR.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Identification, characterization, and expression analysis of auxin response factor (ARF) gene family in Brachypodium distachyon.Funct Integr Genomics. 2018 Nov;18(6):709-724. doi: 10.1007/s10142-018-0622-z. Epub 2018 Jun 20. Funct Integr Genomics. 2018. PMID: 29926224
-
Molecular Characterization and Expression Profiling of NAC Transcription Factors in Brachypodium distachyon L.PLoS One. 2015 Oct 7;10(10):e0139794. doi: 10.1371/journal.pone.0139794. eCollection 2015. PLoS One. 2015. PMID: 26444425 Free PMC article.
-
Genome-Wide Analysis and Expression Profiles of the MYB Genes in Brachypodium distachyon.Plant Cell Physiol. 2017 Oct 1;58(10):1777-1788. doi: 10.1093/pcp/pcx115. Plant Cell Physiol. 2017. PMID: 29016897
-
[Auxin response factors and plant growth and development].Yi Chuan. 2011 Dec;33(12):1335-46. doi: 10.3724/sp.j.1005.2011.01335. Yi Chuan. 2011. PMID: 22207379 Review. Chinese.
-
Genetic dissection of the auxin response network.Nat Plants. 2020 Sep;6(9):1082-1090. doi: 10.1038/s41477-020-0739-7. Epub 2020 Aug 17. Nat Plants. 2020. PMID: 32807951 Review.
Cited by
-
Genome-Wide Identification, Evolution, and Expression Analysis of TPS and TPP Gene Families in Brachypodium distachyon.Plants (Basel). 2019 Sep 23;8(10):362. doi: 10.3390/plants8100362. Plants (Basel). 2019. PMID: 31547557 Free PMC article.
-
MicroRNA and Degradome Profiling Uncover Defense Response of Fraxinus velutina Torr. to Salt Stress.Front Plant Sci. 2022 Apr 1;13:847853. doi: 10.3389/fpls.2022.847853. eCollection 2022. Front Plant Sci. 2022. PMID: 35432418 Free PMC article.
-
Genome-Wide Identification and Expression Analysis of Auxin Response Factor Gene Family in Linum usitatissimum.Int J Mol Sci. 2023 Jul 2;24(13):11006. doi: 10.3390/ijms241311006. Int J Mol Sci. 2023. PMID: 37446183 Free PMC article.
-
Identification of ARF transcription factor gene family and its defense responses to bacterial infection and salicylic acid treatment in sugarcane.Front Microbiol. 2023 Sep 7;14:1257355. doi: 10.3389/fmicb.2023.1257355. eCollection 2023. Front Microbiol. 2023. PMID: 37744907 Free PMC article.
-
Molecular Evolution and Local Root Heterogeneous Expression of the Chenopodium quinoa ARF Genes Provide Insights into the Adaptive Domestication of Crops in Complex Environments.J Mol Evol. 2021 Jun;89(4-5):287-301. doi: 10.1007/s00239-021-10005-5. Epub 2021 Mar 23. J Mol Evol. 2021. PMID: 33755734
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

