High genetic diversity and distinct ancient lineage of Asiatic black bears revealed by non-invasive surveys in the Annapurna Conservation Area, Nepal
- PMID: 30517155
- PMCID: PMC6281213
- DOI: 10.1371/journal.pone.0207662
High genetic diversity and distinct ancient lineage of Asiatic black bears revealed by non-invasive surveys in the Annapurna Conservation Area, Nepal
Abstract
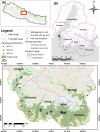
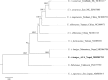
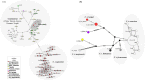
Asiatic black bears (Ursus thibetanus) have a widespread distribution in mountain landscapes, and are considered vulnerable globally, but are low-priority species for conservation in Nepal. Habitat fragmentation, illegal hunting, and human-bear conflict are the major threats to Asiatic black bears across their global range. Having an adequate level of genetic variation in a population helps with adapting to rapidly changing environments, and thus is important for the long-term health of bear populations. Accordingly, we conducted non-invasive surveys of bear populations in the Annapurna Conservation Area (ACA) to elucidate genetic diversity, genetic structure, and the phylogenetic relationship of Asiatic black bears from this region of Nepal to other subspecies. To assess levels of genetic diversity and population genetic structure, we genotyped eight microsatellite loci using 147 samples, identifying 60 individuals in an area of approximately 525 km2. We found that the Asiatic black bear population in the ACA has maintained high levels of genetic diversity (HE = 0.76) as compared to other bear populations from range countries. We did not detect a signature of population substructure among sampling localities and this suggests that animals are moving freely across the landscape within the ACA. We also detected a moderate population size that may increase with the availability of suitable habitat in the ACA, so bear-related conflict should be addressed to ensure the long-term viability of this expanding bear populations. Primers specific to bears were designed to amplify a 675 bp fragment of the mitochondrial control region from the collected samples. Three haplotypes were observed from the entire conservation area. The complete mitochondrial genome (16,771 bp), the first obtained from wild populations of the Himalayan black bear (U. t. laniger), was also sequenced to resolve the phylogenetic relationships of closely related subspecies of Asiatic black bears. The resulting phylogeny indicated that Himalayan black bear populations in Nepal are evolutionary distinct from other known subspecies of Asiatic black bears.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Predicting the potential habitat of bears under a changing climate in Nepal.Environ Monit Assess. 2024 Oct 23;196(11):1097. doi: 10.1007/s10661-024-13253-2. Environ Monit Assess. 2024. PMID: 39443401 Free PMC article.
-
Genetic status of Asiatic black bear (Ursus thibetanus) reintroduced into South Korea based on mitochondrial DNA and microsatellite loci analysis.J Hered. 2011 Mar-Apr;102(2):165-74. doi: 10.1093/jhered/esq121. Epub 2011 Feb 15. J Hered. 2011. PMID: 21325020
-
Genetic structure of the asiatic black bear in Japan using mitochondrial DNA analysis.J Hered. 2009 May-Jun;100(3):297-308. doi: 10.1093/jhered/esn097. Epub 2008 Nov 4. J Hered. 2009. PMID: 18984857
-
Identifying the environmental and anthropogenic causes, distribution, and intensity of human rhesus macaque conflict in Nepal.J Environ Manage. 2022 Aug 15;316:115276. doi: 10.1016/j.jenvman.2022.115276. Epub 2022 May 13. J Environ Manage. 2022. PMID: 35576709 Review.
-
Evaluating hybrid speciation and swamping in wild carnivores with a decision-tree approach.Conserv Biol. 2024 Feb;38(1):e14197. doi: 10.1111/cobi.14197. Epub 2023 Nov 22. Conserv Biol. 2024. PMID: 37811741 Review.
Cited by
-
Predicting the potential habitat of bears under a changing climate in Nepal.Environ Monit Assess. 2024 Oct 23;196(11):1097. doi: 10.1007/s10661-024-13253-2. Environ Monit Assess. 2024. PMID: 39443401 Free PMC article.
-
Asiatic Black Bear-Human Conflict: A Case Study from Guthichaur Rural Municipality, Jumla, Nepal.Animals (Basel). 2024 Apr 17;14(8):1206. doi: 10.3390/ani14081206. Animals (Basel). 2024. PMID: 38672357 Free PMC article.
-
Assembling mitogenome of Himalayan Black Bear (U. t. laniger) from low depth reads and its application in drawing phylogenetic inferences.Sci Rep. 2021 Jan 12;11(1):730. doi: 10.1038/s41598-020-76872-y. Sci Rep. 2021. PMID: 33436634 Free PMC article.
-
Habitat occupancy of sloth bear Melursus ursinus in Chitwan National Park, Nepal.Ecol Evol. 2022 Mar 6;12(3):e8699. doi: 10.1002/ece3.8699. eCollection 2022 Mar. Ecol Evol. 2022. PMID: 35342572 Free PMC article.
References
-
- Garshelis D, Steinmetz R. Ursus thibetanus (errata version published in 2017). The IUCN Red List of Threatened Species. 2016:e.T22824A114252336. Available from: 10.2305/IUCN.UK.2016-3.RLTS.T22824A45034242.en. Cited 24 February 2018. - DOI
-
- Ohnishi N, Uno R, Ishibashi Y, Tamate HB, Oi T. The influence of climatic oscillations during the Quaternary Era on the genetic structure of Asian black bears in Japan. Heredity (Edinb). 2009;102(6):579–589. 10.1038/hdy.2009.28 - DOI - PubMed
-
- Wu J, Kohno N, Mano S, Fukumoto Y, Tanabe H, Hasegawa M, et al. Phylogeographic and demographic analysis of the Asian black bear (Ursus thibetanus) based on mitochondrial DNA. PLoS ONE. 2015;10(9):e0136398 10.1371/journal.pone.0136398 - DOI - PMC - PubMed
-
- Heptner VG, Naumov NP. Mammals of the Soviet Union. Vol. II Part 1a, Sirenia and Carnivora (Sea cows; Wolves and Bears). Washington DC: Smithsonian Institution libraries and National Science Foundation; 1998.
-
- Hwang DS, Ki JS, Jeong DH, Kim BH, Lee BK, Han SH, et al. A comprehensive analysis of three Asiatic black bear mitochondrial genomes (subspecies ussuricus, formosanus and mupinensis), with emphasis on the complete mtDNA sequence of Ursus thibetanus ussuricus (Ursidae). Mitochondrial DNA. 2008;19(4):418–429. 10.1080/19401730802389525 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

