Advances in multiple omics of natural-killer/T cell lymphoma
- PMID: 30514323
- PMCID: PMC6280527
- DOI: 10.1186/s13045-018-0678-1
Advances in multiple omics of natural-killer/T cell lymphoma
Abstract
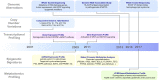
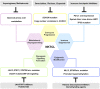
Natural-killer/T cell lymphoma (NKTCL) represents the most common subtype of extranodal lymphoma with aggressive clinical behavior. Prevalent in Asians and South Americans, the pathogenesis of NKTCL remains to be fully elucidated. Using system biology techniques including genomics, transcriptomics, epigenomics, and metabolomics, novel biomarkers and therapeutic targets have been revealed in NKTCL. Whole-exome sequencing studies identify recurrent somatic gene mutations, involving RNA helicases, tumor suppressors, JAK-STAT pathway molecules, and epigenetic modifiers. Another genome-wide association study reports that single nucleotide polymorphisms mapping to the class II MHC region on chromosome 6 contribute to lymphomagenesis. Alterations of oncogenic signaling pathways janus kinase-signal transducer and activator of transcription (JAK-STAT), nuclear factor-κB (NF-κB), mitogen-activated protein kinase (MAPK), WNT, and NOTCH, as well as epigenetic dysregulation of microRNA and long non-coding RNAs, are also frequently observed in NKTCL. As for metabolomic profiling, abnormal amino acids metabolism plays an important role on disease progression of NKTCL. Of note, through targeting multiple omics aberrations, clinical outcome of NKTCL patients has been significantly improved by asparaginase-based regimens, immune checkpoints inhibitors, and histone deacetylation inhibitors. Future investigations will be emphasized on molecular classification of NKTCL using integrated analysis of system biology, so as to optimize targeted therapeutic strategies of NKTCL in the era of precision medicine.
Keywords: Epigenomics; Genomics; Metabolomics; Natural-killer/T cell lymphomas; Targeted therapy; Transcriptomics.
Conflict of interest statement
Authors’ information
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
What we should know about natural killer/T-cell lymphomas.Hematol Oncol. 2019 Jun;37 Suppl 1:75-81. doi: 10.1002/hon.2588. Hematol Oncol. 2019. PMID: 31187536 Review.
-
SLC1A1 mediated glutamine addiction and contributed to natural killer T-cell lymphoma progression with immunotherapeutic potential.EBioMedicine. 2021 Oct;72:103614. doi: 10.1016/j.ebiom.2021.103614. Epub 2021 Oct 7. EBioMedicine. 2021. PMID: 34628354 Free PMC article.
-
The Genomics and Molecular Biology of Natural Killer/T-Cell Lymphoma: Opportunities for Translation.Int J Mol Sci. 2018 Jun 30;19(7):1931. doi: 10.3390/ijms19071931. Int J Mol Sci. 2018. PMID: 29966370 Free PMC article. Review.
-
CD56-Negative Extranodal Natural Killer/T-Cell Lymphoma: A Retrospective Study in 443 Patients Treated by Chemotherapy With or Without Asparaginase.Front Immunol. 2022 Mar 17;13:829366. doi: 10.3389/fimmu.2022.829366. eCollection 2022. Front Immunol. 2022. PMID: 35371002 Free PMC article.
-
Genetic risk of extranodal natural killer T-cell lymphoma: a genome-wide association study.Lancet Oncol. 2016 Sep;17(9):1240-7. doi: 10.1016/S1470-2045(16)30148-6. Epub 2016 Jul 25. Lancet Oncol. 2016. PMID: 27470079 Free PMC article.
Cited by
-
Novel target and treatment agents for natural killer/T-cell lymphoma.J Hematol Oncol. 2023 Jul 22;16(1):78. doi: 10.1186/s13045-023-01483-9. J Hematol Oncol. 2023. PMID: 37480137 Free PMC article. Review.
-
Analysis of mutation profiles in extranodal NK/T-cell lymphoma: clinical and prognostic correlations.Ann Hematol. 2024 Aug;103(8):2917-2930. doi: 10.1007/s00277-024-05698-9. Epub 2024 Apr 27. Ann Hematol. 2024. PMID: 38671297
-
EBV and the Pathogenesis of NK/T Cell Lymphoma.Cancers (Basel). 2021 Mar 19;13(6):1414. doi: 10.3390/cancers13061414. Cancers (Basel). 2021. PMID: 33808787 Free PMC article. Review.
-
Intravascular NK/T-Cell Lymphoma: What We Know about This Diagnostically Challenging, Aggressive Disease.Cancers (Basel). 2022 Nov 6;14(21):5458. doi: 10.3390/cancers14215458. Cancers (Basel). 2022. PMID: 36358876 Free PMC article. Review.
-
Intestinal natural killer/T-cell lymphoma presenting as a pancreatic head space-occupying lesion: A case report.World J Gastrointest Oncol. 2023 Jan 15;15(1):195-204. doi: 10.4251/wjgo.v15.i1.195. World J Gastrointest Oncol. 2023. PMID: 36684049 Free PMC article.
References
-
- Hong M, Lee T, Young Kang S, Kim SJ, Kim W, Ko YH. Nasal-type NK/T-cell lymphomas are more frequently T rather than NK lineage based on T-cell receptor gene, RNA, and protein studies: lineage does not predict clinical behavior. Mod Pathol. 2016;29(5):430–443. doi: 10.1038/modpathol.2016.47. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

