In Cellulo Protein-mRNA Interaction Assay to Determine the Action of G-Quadruplex-Binding Molecules
- PMID: 30501034
- PMCID: PMC6321085
- DOI: 10.3390/molecules23123124
In Cellulo Protein-mRNA Interaction Assay to Determine the Action of G-Quadruplex-Binding Molecules
Abstract
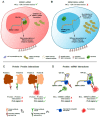
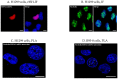
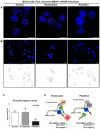
Protein-RNA interactions (PRIs) control pivotal steps in RNA biogenesis, regulate multiple physiological and pathological cellular networks, and are emerging as important drug targets. However, targeting of specific protein-RNA interactions for therapeutic developments is still poorly advanced. Studies and manipulation of these interactions are technically challenging and in vitro drug screening assays are often hampered due to the complexity of RNA structures. The binding of nucleolin (NCL) to a G-quadruplex (G4) structure in the messenger RNA (mRNA) of the Epstein-Barr virus (EBV)-encoded EBNA1 has emerged as an interesting therapeutic target to interfere with immune evasion of EBV-associated cancers. Using the NCL-EBNA1 mRNA interaction as a model, we describe a quantitative proximity ligation assay (PLA)-based in cellulo approach to determine the structure activity relationship of small chemical G4 ligands. Our results show how different G4 ligands have different effects on NCL binding to G4 of the EBNA1 mRNA and highlight the importance of in-cellulo screening assays for targeting RNA structure-dependent interactions.
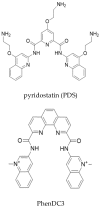
Keywords: EBNA1; Epstein-Barr virus (EBV); G-quadruplexes; PhenDC3; protein-mRNA interactions; pyridostatin; structure-activity relationship.
Conflict of interest statement
The authors declare no commercial or financial conflict of interest.
Figures
Similar articles
-
Type I arginine methyltransferases are intervention points to unveil the oncogenic Epstein-Barr virus to the immune system.Nucleic Acids Res. 2022 Nov 11;50(20):11799-11819. doi: 10.1093/nar/gkac915. Nucleic Acids Res. 2022. PMID: 36350639 Free PMC article.
-
Novel cationic bis(acylhydrazones) as modulators of Epstein-Barr virus immune evasion acting through disruption of interaction between nucleolin and G-quadruplexes of EBNA1 mRNA.Eur J Med Chem. 2019 Sep 15;178:13-29. doi: 10.1016/j.ejmech.2019.05.042. Epub 2019 May 23. Eur J Med Chem. 2019. PMID: 31173968
-
Nucleolin directly mediates Epstein-Barr virus immune evasion through binding to G-quadruplexes of EBNA1 mRNA.Nat Commun. 2017 Jul 7;8:16043. doi: 10.1038/ncomms16043. Nat Commun. 2017. PMID: 28685753 Free PMC article.
-
Epstein-Barr virus-encoded EBNA1 and ZEBRA: targets for therapeutic strategies against EBV-carrying cancers.J Pathol. 2015 Jan;235(2):334-41. doi: 10.1002/path.4431. J Pathol. 2015. PMID: 25186125 Review.
-
Targeting the RNA G-Quadruplex and Protein Interactome for Antiviral Therapy.J Med Chem. 2022 Aug 11;65(15):10161-10182. doi: 10.1021/acs.jmedchem.2c00649. Epub 2022 Jul 21. J Med Chem. 2022. PMID: 35862260 Review.
Cited by
-
Sneaking Out for Happy Hour: Yeast-Based Approaches to Explore and Modulate Immune Response and Immune Evasion.Genes (Basel). 2019 Aug 31;10(9):667. doi: 10.3390/genes10090667. Genes (Basel). 2019. PMID: 31480411 Free PMC article. Review.
-
G-Quadruplex Targeting in the Fight against Viruses: An Update.Int J Mol Sci. 2021 Oct 12;22(20):10984. doi: 10.3390/ijms222010984. Int J Mol Sci. 2021. PMID: 34681641 Free PMC article. Review.
-
Phosphorylation of cytochrome c at tyrosine 48 finely regulates its binding to the histone chaperone SET/TAF-Iβ in the nucleus.Protein Sci. 2024 Dec;33(12):e5213. doi: 10.1002/pro.5213. Protein Sci. 2024. PMID: 39548742 Free PMC article.
-
Can G-quadruplex become a promising target in HBV therapy?Front Immunol. 2022 Dec 15;13:1091873. doi: 10.3389/fimmu.2022.1091873. eCollection 2022. Front Immunol. 2022. PMID: 36591216 Free PMC article. Review.
-
Type I arginine methyltransferases are intervention points to unveil the oncogenic Epstein-Barr virus to the immune system.Nucleic Acids Res. 2022 Nov 11;50(20):11799-11819. doi: 10.1093/nar/gkac915. Nucleic Acids Res. 2022. PMID: 36350639 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

