Kinetic and catalytic properties of M.HpyAXVII, a phase-variable DNA methyltransferase from Helicobacter pylori
- PMID: 30478171
- PMCID: PMC6341398
- DOI: 10.1074/jbc.RA118.003769
Kinetic and catalytic properties of M.HpyAXVII, a phase-variable DNA methyltransferase from Helicobacter pylori
Erratum in
-
Correction: Kinetic and catalytic properties of M.HpyAXVII, a phase-variable DNA methyltransferase from Helicobacter pylori.J Biol Chem. 2019 Aug 30;294(35):13199. doi: 10.1074/jbc.AAC119.010443. J Biol Chem. 2019. PMID: 31471381 Free PMC article. No abstract available.
Abstract
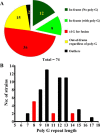
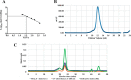
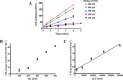
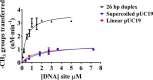
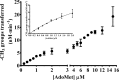
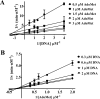
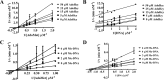
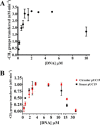
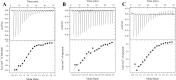
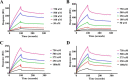
The bacterium Helicobacter pylori is one of the most common infectious agents found in the human stomach. H. pylori has an unusually large number of DNA methyltransferases (MTases), prompting speculation that they may be involved in the cancerization of epithelial cells. The mod-4a/4b locus, consisting of the hp1369 and hp1370 ORFs, encodes for a truncated and inactive MTase in H. pylori strain 26695. However, slipped-strand synthesis within the phase-variable polyguanine tract in hp1369 results in expression of an active HP1369-1370 fusion N6-adenine methyltransferase, designated M.HpyAXVII. Sequence analysis of the mod-4a/4b locus across 74 H. pylori strain genomes has provided insights into the regulation of M.HpyAXVII expression. To better understand the role of M.HpyAXVII in the H. pylori biology, here we cloned and overexpressed the hp1369-70 fusion construct in Escherichia coli BL21(DE3) cells. Results from size-exclusion chromatography and multi-angle light scattering (MALS) analyses suggested that M.HpyAXVII exists as a dimer in solution. Kinetic studies, including product and substrate inhibition analyses, initial velocity dependence between substrates, and isotope partitioning, suggested that M.HpyAXVII catalyzes DNA methylation in an ordered Bi Bi mechanism in which the AdoMet binding precedes DNA binding and AdoMet's methyl group is then transferred to an adenine within the DNA recognition sequence. Altering the highly conserved catalytic motif (DPP(Y/F)) as well as the AdoMet-binding motif (FXGXG) by site-directed mutagenesis abolished the catalytic activity of M.HpyAXVII. These results provide insights into the enzyme kinetic mechanism of M.HpyAXVII. We propose that AdoMet binding conformationally "primes" the enzyme for DNA binding.
Keywords: DNA binding; DNA methyltransferase; M.HpyAXVII; S-adenosylmethionine (AdoMet); allosteric regulation; bioinformatics; enzyme kinetics; enzyme mechanism; ordered Bi Bi mechanism; phase variation.
© 2019 Prasad et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


Similar articles
-
Functional analysis of an acid adaptive DNA adenine methyltransferase from Helicobacter pylori 26695.PLoS One. 2011 Feb 9;6(2):e16810. doi: 10.1371/journal.pone.0016810. PLoS One. 2011. PMID: 21347417 Free PMC article.
-
A nucleotide insertion between two adjacent methyltransferases in Helicobacter pylori results in a bifunctional DNA methyltransferase.Biochem J. 2011 Feb 1;433(3):487-95. doi: 10.1042/BJ20101668. Biochem J. 2011. PMID: 21110832
-
Functional analysis of the M.HpyAIV DNA methyltransferase of Helicobacter pylori.J Bacteriol. 2007 Dec;189(24):8914-21. doi: 10.1128/JB.00108-07. Epub 2007 Oct 5. J Bacteriol. 2007. PMID: 17921292 Free PMC article.
-
The Helicobacter pylori Methylome: Roles in Gene Regulation and Virulence.Curr Top Microbiol Immunol. 2017;400:105-127. doi: 10.1007/978-3-319-50520-6_5. Curr Top Microbiol Immunol. 2017. PMID: 28124151 Review.
-
DNA methyltransferases: mechanistic models derived from kinetic analysis.Crit Rev Biochem Mol Biol. 2012 Mar-Apr;47(2):97-193. doi: 10.3109/10409238.2011.620942. Epub 2012 Jan 20. Crit Rev Biochem Mol Biol. 2012. PMID: 22260147 Review.
Cited by
-
Beta class amino methyltransferases from bacteria to humans: evolution and structural consequences.Nucleic Acids Res. 2020 Oct 9;48(18):10034-10044. doi: 10.1093/nar/gkaa446. Nucleic Acids Res. 2020. PMID: 32453412 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

